
Rice Science ›› 2022, Vol. 29 ›› Issue (6): 545-558.DOI: 10.1016/j.rsci.2022.04.002
• Research Paper • Previous Articles Next Articles
Blaise Pascal Muvunyi1, Lu Xiang1, Zhan Junhui1, He Sang1( ), Ye Guoyou1,2(
), Ye Guoyou1,2( )
)
Received:2021-12-09
Accepted:2022-04-24
Online:2022-11-28
Published:2022-09-09
Contact:
He Sang, Ye Guoyou
Blaise Pascal Muvunyi, Lu Xiang, Zhan Junhui, He Sang, Ye Guoyou. Identification of Potential Zinc Deficiency Responsive Genes and Regulatory Pathways in Rice by Weighted Gene Co-expression Network Analysis[J]. Rice Science, 2022, 29(6): 545-558.
Add to citation manager EndNote|Ris|BibTeX
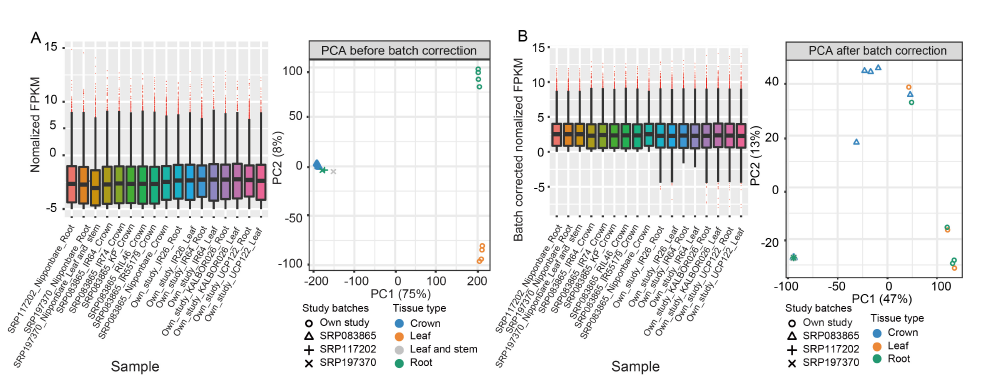
Fig. 1. Box plot and principal component analysis (PCA) using normalized fragments per kilobase of transcript per million mapped fragments (FPKM) before (A) and after (B) batch correction in 17 samples. Samples with SRP197370, SRP083865 and SRP117202 were from Zeng et al (2019b), Nanda et al (2017) and Lu et al (2021), respectively. Only samples in the first two PCA axis were shown.
| Module label | No. of genes | Top GO term (FDR < 0.05) | Top KEGG pathway (FDR < 0.05) | Validated gene a |
|---|---|---|---|---|
| A | 6 304 | Multicellular organism development; multicellular organismal process, etc | Interactions of soluble N-ethylmaleimide-sensitive factor attachment protein receptors in vesicular transport; autophagy, etc | OsZIFL2, OsYSL18, OsZIP4, OsFRDL1, OsDMAS1, OsNAS1, OsYSL4, ONAC036, OsbZIP48, OsbZIP50, OsZIFL3, OsZIP8, OsFER1, OsZIFL5, OsMT1a |
| B | 4 316 | Terpenoid metabolic process; isoprenoid metabolic process, etc | Photosynthesis-antenna proteins; cutin, suberine and wax biosynthesis, etc | OsNRAMP4, OsYSL14, OsZIP3, OsMTP1, OsGT1, OsOPT4, OsVIT2, FER2 |
| C | 1 639 | DNA metabolic process; purine nucleotide biosynthetic process, etc | Ribosome; tyrosine metabolism, etc | OsYSL9, OsNAAT4, OsNAS3, OsZIP9 |
| D | 1 167 | Intracellular signal transduction; regulation of transcription, etc | Biosynthesis of amino acids, phenylalanine, tyrosine, tryptophan and phenylpropanoid, etc | OsSAM2, OsNAC4, OsYSL12, OsZIP5, OsZIFL7 |
| E | 1 049 | Nucleic acid metabolic process; RNA metabolic process | Alpha-linolenic acid metabolism; plant-pathogen interaction | OsZIP2, OsTOM1, OsNRAMP7 |
| F | 909 | Cellular response to stimulus | Peroxisome; MAPK signaling pathway-plant, etc | |
| G | 877 | No significant GO annotation found | Nucleotide excision repair; selenocompound metabolism, etc | OsNAAT1, OsYSL6, OsSAMS1, OsZP7, OsZIP10 |
| H | 696 | No significant GO annotation found | Ether lipid metabolism; photosynthesis, etc | OsOPT, OsHMA2, OsHMA3 |
| I | 667 | Cellular component organization; cellular component organization or biogenesis | Phagosome; phenylpropanoid biosynthesis; MAPK signaling pathway-plant; nicotinate and nicotinamide metabolism | OsYSL10 |
| J | 461 | Organic substance transport, etc | Nitrogen metabolism; amino sugar and nucleotide sugar metabolism, etc | OsZIP1 |
| K | 367 | Cellular protein metabolic process; protein metabolic process, etc | Amino acid biosynthesis; valine, leucine, and isoleucine degradation; propanoate metabolism, etc | OsNRAMP6, OsIRO2, OsZIFL12 |
| L | 240 | No significant GO annotation found | Base excision repair; mRNA surveillance pathway, etc | |
| M | 123 | Aerobic respiration; cellular respiration | No significant KEGG annotations found | |
| N | 112 | Cell part; cell | RNA transport; mRNA surveillance pathway | |
| O | 94 | No significant GO annotation found | Carbon fixation in photosynthetic organisms; ribosome | |
| P | 61 | No significant GO annotation found | Zeatin biosynthesis |
Table 1. Modules’ functional annotation and distribution of functionally validated Zn deficiency responsive genes in detected modules.
| Module label | No. of genes | Top GO term (FDR < 0.05) | Top KEGG pathway (FDR < 0.05) | Validated gene a |
|---|---|---|---|---|
| A | 6 304 | Multicellular organism development; multicellular organismal process, etc | Interactions of soluble N-ethylmaleimide-sensitive factor attachment protein receptors in vesicular transport; autophagy, etc | OsZIFL2, OsYSL18, OsZIP4, OsFRDL1, OsDMAS1, OsNAS1, OsYSL4, ONAC036, OsbZIP48, OsbZIP50, OsZIFL3, OsZIP8, OsFER1, OsZIFL5, OsMT1a |
| B | 4 316 | Terpenoid metabolic process; isoprenoid metabolic process, etc | Photosynthesis-antenna proteins; cutin, suberine and wax biosynthesis, etc | OsNRAMP4, OsYSL14, OsZIP3, OsMTP1, OsGT1, OsOPT4, OsVIT2, FER2 |
| C | 1 639 | DNA metabolic process; purine nucleotide biosynthetic process, etc | Ribosome; tyrosine metabolism, etc | OsYSL9, OsNAAT4, OsNAS3, OsZIP9 |
| D | 1 167 | Intracellular signal transduction; regulation of transcription, etc | Biosynthesis of amino acids, phenylalanine, tyrosine, tryptophan and phenylpropanoid, etc | OsSAM2, OsNAC4, OsYSL12, OsZIP5, OsZIFL7 |
| E | 1 049 | Nucleic acid metabolic process; RNA metabolic process | Alpha-linolenic acid metabolism; plant-pathogen interaction | OsZIP2, OsTOM1, OsNRAMP7 |
| F | 909 | Cellular response to stimulus | Peroxisome; MAPK signaling pathway-plant, etc | |
| G | 877 | No significant GO annotation found | Nucleotide excision repair; selenocompound metabolism, etc | OsNAAT1, OsYSL6, OsSAMS1, OsZP7, OsZIP10 |
| H | 696 | No significant GO annotation found | Ether lipid metabolism; photosynthesis, etc | OsOPT, OsHMA2, OsHMA3 |
| I | 667 | Cellular component organization; cellular component organization or biogenesis | Phagosome; phenylpropanoid biosynthesis; MAPK signaling pathway-plant; nicotinate and nicotinamide metabolism | OsYSL10 |
| J | 461 | Organic substance transport, etc | Nitrogen metabolism; amino sugar and nucleotide sugar metabolism, etc | OsZIP1 |
| K | 367 | Cellular protein metabolic process; protein metabolic process, etc | Amino acid biosynthesis; valine, leucine, and isoleucine degradation; propanoate metabolism, etc | OsNRAMP6, OsIRO2, OsZIFL12 |
| L | 240 | No significant GO annotation found | Base excision repair; mRNA surveillance pathway, etc | |
| M | 123 | Aerobic respiration; cellular respiration | No significant KEGG annotations found | |
| N | 112 | Cell part; cell | RNA transport; mRNA surveillance pathway | |
| O | 94 | No significant GO annotation found | Carbon fixation in photosynthetic organisms; ribosome | |
| P | 61 | No significant GO annotation found | Zeatin biosynthesis |
| GO term | Term description | False discovery rate | Gene entry |
|---|---|---|---|
| GO:0022891 | Substrate-specific transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0022892 | Substrate-specific transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0022857 | Transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0015075 | Ion transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5 |
| GO:0031224 | Intrinsic to membrane | 0.00022 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0016021 | Integral to membrane | 0.00022 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0006812 | Cation transport | 0.00032 | OsHMA1, OsZIP5, OsZIP8, OsZIP9, OsA2 |
| GO:0006811 | Ion transport | 0.00034 | OsHMA1, OsZIP5, OsZIP8, OsZIP9, OsA2 |
| GO:0005215 | Transporter activity | 0.00063 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0044425 | Membrane part | 0.00063 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0044281 | Small molecule metabolic process | 0.00182 | OsLAC29, OsHMA1, OsIGPS, OsNAS3, OsNAAT1, OsA2 |
| GO:0006810 | Transport | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0051234 | Establishment of localization | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0051179 | Localization | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0016020 | Membrane | 0.02200 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
Table 2. Gene ontology (GO) annotations significantly enriched by conserved and up-regulated modular genes.
| GO term | Term description | False discovery rate | Gene entry |
|---|---|---|---|
| GO:0022891 | Substrate-specific transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0022892 | Substrate-specific transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0022857 | Transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0015075 | Ion transmembrane transporter activity | 0.00010 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5 |
| GO:0031224 | Intrinsic to membrane | 0.00022 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0016021 | Integral to membrane | 0.00022 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0006812 | Cation transport | 0.00032 | OsHMA1, OsZIP5, OsZIP8, OsZIP9, OsA2 |
| GO:0006811 | Ion transport | 0.00034 | OsHMA1, OsZIP5, OsZIP8, OsZIP9, OsA2 |
| GO:0005215 | Transporter activity | 0.00063 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0044425 | Membrane part | 0.00063 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0044281 | Small molecule metabolic process | 0.00182 | OsLAC29, OsHMA1, OsIGPS, OsNAS3, OsNAAT1, OsA2 |
| GO:0006810 | Transport | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0051234 | Establishment of localization | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0051179 | Localization | 0.01100 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
| GO:0016020 | Membrane | 0.02200 | OsHMA1, OsZIP8, OsZIP9, OsMST4, OsZIP5, OsA2 |
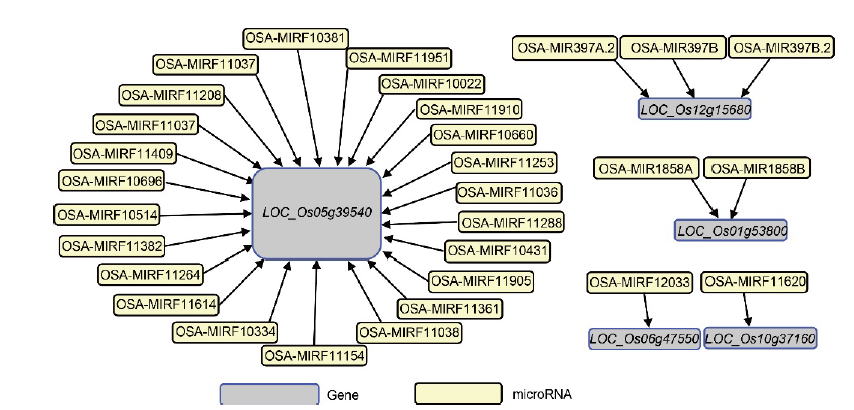
Fig. 2. miRNAs targeting the identified conserved differentially expressed modular genes. miRNAs were retrieved from http://structuralbiology.cau.edu.cn/PlantGSEA/database (Yi et al, 2013).
| Gene ID | Gene name | Gene function a | Module |
|---|---|---|---|
| LOC_Os12g41950 | OsARF6b b | Regulates grain Fe accumulation by controlling rice crown root formation via cytokinin and auxin signaling | A |
| LOC_ Os07g09340 | OsA2 c | Positively regulates net flux of NO3-, nitrogen concentration; improves grain yield | A |
| LOC_Os09g28210 | OsbHLH120 b | Regulates root thickness and length in upland rice | B |
| LOC_Os07g48560 | OsWOX11 b | Enhances crown root development, drought stress tolerance, and potassium deficiency tolerance; regulates cytokinin-signaling pathway | D |
| LOC_Os11g45740 | OsJAmyb b | Improves root growth in seedlings, tolerances to blast diseases and salt stress | D |
| LOC_Os11g03300 | NAC122 b | Enhances grain yield, drought and disease tolerances | D |
| LOC_Os08g09690 | HAP2A b | Positively regulates drought stress tolerance | D |
| LOC_Os06g03670 | OsDREB1C b | Involves in stress response | E |
| LOC_Os12g36850 | RPR10b c | Involves in root biotic and abiotic stress responses, potentially, under the jasmonic acid signaling pathway | H |
| LOC_Os03g11900 | OsMST4 c | Functions in monosaccharides transport during grain filling period | G |
| LOC_Os04g23910 | OsMADS25 c | Regulates root system architecture via auxin signaling | G |
| LOC_Os05g03884 | Oskn2 b | Involves in embryo, shoot, and flower development | G |
| LOC_Os02g57490 | OsDH1 b | Regulates glume development | G |
| LOC_Os03g48270 | OsCDPK9 d | Confers drought stress resistance and spikelet fertility | G |
| LOC_Os05g49140 | OsMPK7 d | Involves in drought stress tolerance | G |
Table 3. Known functions of regulatory genes interacting with conserved differentially expressed modular genes.
| Gene ID | Gene name | Gene function a | Module |
|---|---|---|---|
| LOC_Os12g41950 | OsARF6b b | Regulates grain Fe accumulation by controlling rice crown root formation via cytokinin and auxin signaling | A |
| LOC_ Os07g09340 | OsA2 c | Positively regulates net flux of NO3-, nitrogen concentration; improves grain yield | A |
| LOC_Os09g28210 | OsbHLH120 b | Regulates root thickness and length in upland rice | B |
| LOC_Os07g48560 | OsWOX11 b | Enhances crown root development, drought stress tolerance, and potassium deficiency tolerance; regulates cytokinin-signaling pathway | D |
| LOC_Os11g45740 | OsJAmyb b | Improves root growth in seedlings, tolerances to blast diseases and salt stress | D |
| LOC_Os11g03300 | NAC122 b | Enhances grain yield, drought and disease tolerances | D |
| LOC_Os08g09690 | HAP2A b | Positively regulates drought stress tolerance | D |
| LOC_Os06g03670 | OsDREB1C b | Involves in stress response | E |
| LOC_Os12g36850 | RPR10b c | Involves in root biotic and abiotic stress responses, potentially, under the jasmonic acid signaling pathway | H |
| LOC_Os03g11900 | OsMST4 c | Functions in monosaccharides transport during grain filling period | G |
| LOC_Os04g23910 | OsMADS25 c | Regulates root system architecture via auxin signaling | G |
| LOC_Os05g03884 | Oskn2 b | Involves in embryo, shoot, and flower development | G |
| LOC_Os02g57490 | OsDH1 b | Regulates glume development | G |
| LOC_Os03g48270 | OsCDPK9 d | Confers drought stress resistance and spikelet fertility | G |
| LOC_Os05g49140 | OsMPK7 d | Involves in drought stress tolerance | G |
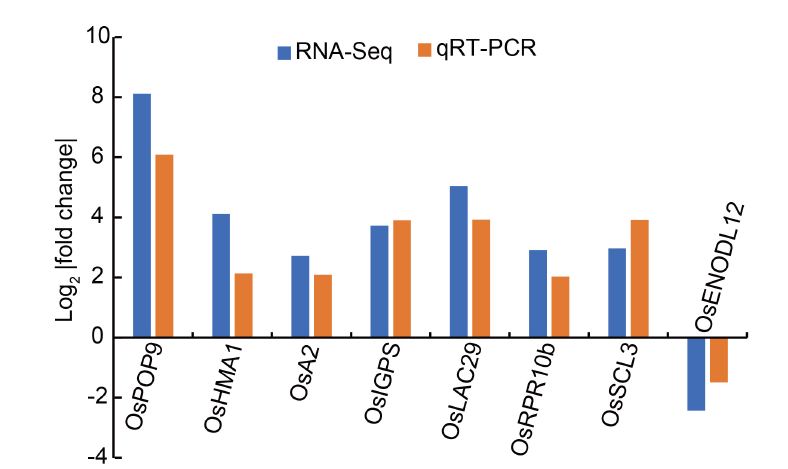
Fig. 3. qRT-PCR validation of RNA-Seq expression of conserved differentially expressed modular genes. RNA-Seq fold changes, which have been reported for root (Table S5) and crown (Table S7) in Nanda et al (2017) and Zeng et al (2019b), respectively, were compared with the obtained qRT-PCR based fold change for the same tissues. During qRT-PCR, expression levels under the control conditions (Zn supply) were normalized to 1 for both root and crown tissues. ACTN-1 was used as an internal reference. The Pearson correlation indicated a positive significant (P < 0.05) correlation (R2 = 0.87) between fold changes obtained based on RNA-Seq and qRT-PCR analyses.
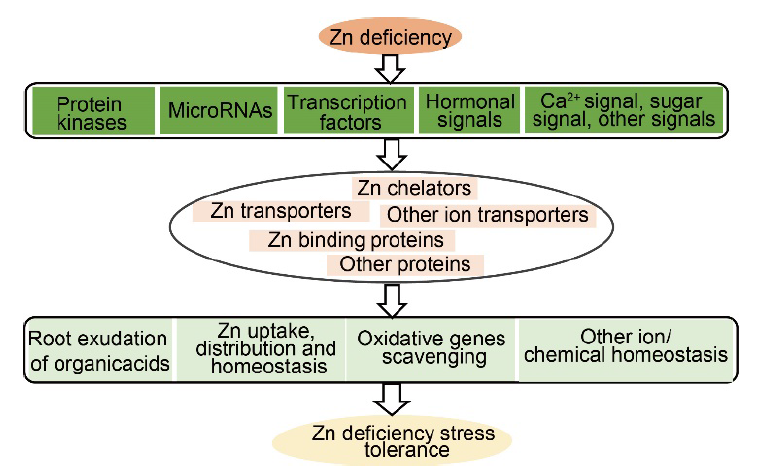
Fig. 4. A theoretical model for signaling pathways underlying Zn deficiency responses in rice. Zn deficiency stress first activates regulatory proteins, such as miRNAs, protein kinases, and transcription factors that modulate various hormonal stress responses, sugar transports, and morphogenetic events. Following that, regulatory proteins induce the expression of the downstream Zn responsive genes, including the small-molecule metabolic process genes, Zn transporter genes, transmembrane transporter genes, metal binding and reactive oxygen species scavenging genes, ion or chemical transporter genes and sugar transporter genes.
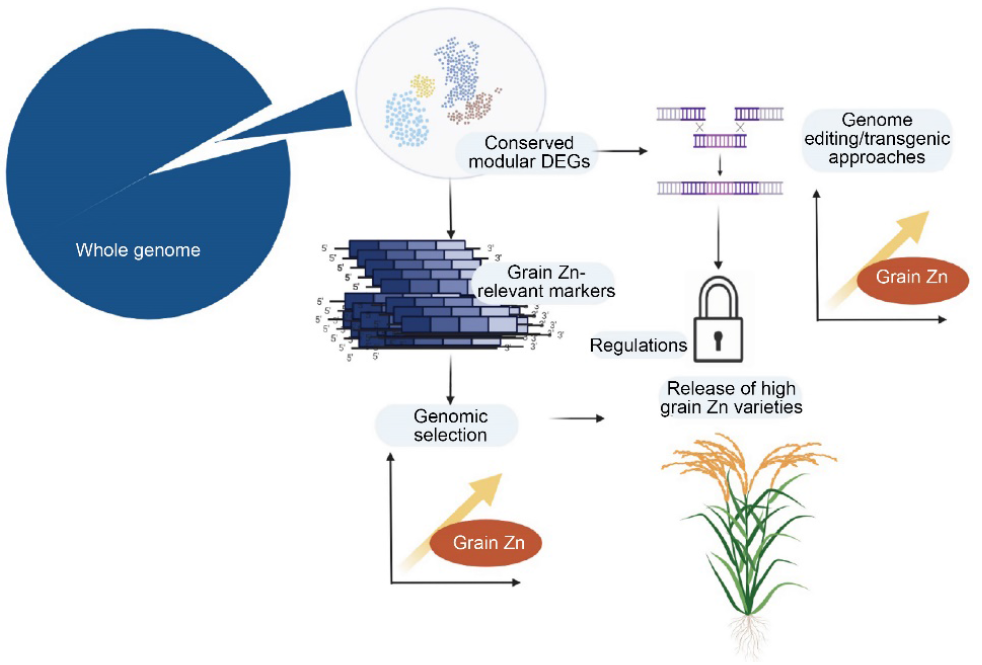
Fig. 5. An illustration of potential use of conserved modular differentially expressed genes (DEGs) in breeding schemes for high grain Zn. Co-expression network analysis allows identification of modules of co-expressed genes. By intersecting modular genes with DEGs reported in different studies on Zn deficiency stress, conserved modular DEGs are obtained. In the next stage, grain Zn-relevant markers can be obtained by selecting features in the proximities of conserved modular DEGs. Finally, the functional markers identified can be deployed in genomic selection models to improve prediction accuracy of high grain Zn lines. Besides genomic selection-based methods, if functionally validated, conserved modular DEGs could also facilitate the development of high-grain Zn via genome editing or transgenic methods. However, food regulations in most developing countries are likely to obstruct delivery of genome-edited or genetically modified biofortified crops.
| [1] | Ahmadi N, Cao T V, Frouin J, Norton G J, Price A H. 2021. Genomic prediction of arsenic tolerance and grain yield in rice: Contribution of trait-specific markers and multi-environment models. Rice Sci, 28(3): 268-278. |
| [2] |
Akhtar S, Das J K, Ismail T, Wahid M, Saeed W, Bhutta Z A. 2021. Nutritional perspectives for the prevention and mitigation of COVID-19. Nutr Rev, 79(3): 289-300.
PMID |
| [3] |
Ariel F D, Manavella P A, Dezar C A, Chan R L. 2007. The true story of the HD-Zip family. Trends Plant Sci, 12(9): 419-426.
PMID |
| [4] | Azodi C B, Pardo J, VanBuren R, de Los Campos G, Shiu S H. 2020. Transcriptome-based prediction of complex traits in maize. Plant Cell, 32(1): 139-151. |
| [5] |
Balyan S, Kumar M, Mutum R D, Raghuvanshi U, Agarwal P, Mathur S, Raghuvanshi S. 2017. Identification of miRNA- mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci Rep, 7(1): 15446.
PMID |
| [6] |
Banakar R, Alvarez Fernandez A, Díaz-Benito P, Abadia J, Capell T, Christou P. 2017. Phytosiderophores determine thresholds for iron and zinc accumulation in biofortified rice endosperm while inhibiting the accumulation of cadmium. J Exp Bot, 68(17): 4983-4995.
PMID |
| [7] | Bandyopadhyay T, Mehra P, Hairat S, Giri J. 2017. Morpho- physiological and transcriptome profiling reveal novel zinc deficiency-responsive genes in rice. Funct Integr Genomics, 17(5): 565-581. |
| [8] | Black M M. 1998. Zinc deficiency and child development. Am J Clin Nutr, 68: 464S-469S. |
| [9] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15): 2114-2120.
PMID |
| [10] | Chen X Y, Mei Q, Liang W F, Sun J, Wang X M, Zhou J, Wang J M, Zhou Y H, Zheng B S, Yang Y, Chen J P. 2020. Gene mapping, genome-wide transcriptome analysis, and WGCNA reveals the molecular mechanism for triggering programmed cell death in rice mutant pir1. Plants, 9(11): 1607. |
| [11] | Cheng S F, Zhou D X, Zhao Y. 2016. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal Behav, 11(2): e1130198. |
| [12] | Childs K L, Davidson R M, Buell C R. 2011. Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS One, 6(7): e22196. |
| [13] |
Cline M S, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico A R, Vailaya A, Wang P L, Adler A, Conklin B R, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner G J, Idker T, Bader G D. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc, 2: 2366-2382.
PMID |
| [14] | Dardenne M. 2002. Zinc and immune function. Eur J Clin Nutr, 56: S20-S23. |
| [15] |
de Los Reyes B G, Mohanty B, Yun S J, Park M R, Lee D Y. 2015. Upstream regulatory architecture of rice genes: Summarizing the baseline towards genus-wide comparative analysis of regulatory networks and allele mining. Rice, 8: 14.
PMID |
| [16] | Du Z, Zhou X, Ling Y, Zhang Z H, Su Z. 2010. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res, 38: W64-W70. |
| [17] | Gratz R, Manishankar P, Ivanov R, Köster P, Mohr I, Trofimov K, Steinhorst L, Meiser J, Mai H J, Drerup M, Arendt S, Holtkamp M, Karst U, Kudla J, Bauer P, Brumbarova T. 2019. CIPK11- dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Dev Cell, 48(5): 726-740. |
| [18] |
Guo R, Dhliwayo T, Mageto E K, Palacios-Rojas N, Lee M, Yu D S, Ruan Y Y, Zhang A, San Vicente F, Olsen M, Crossa J, Prasanna B M, Zhang L J, Zhang X C. 2020. Genomic prediction of kernel zinc concentration in multiple maize populations using genotyping-by-sequencing and repeat amplification sequencing markers. Front Plant Sci, 11: 534.
PMID |
| [19] | Hajiboland R. 2012. Effect of micronutrient deficiencies on plants stress responses. In: Ahmad P, Prasad M. Abiotic Stress Responses in Plants. New York, USA: Springer: 283-329. |
| [20] |
Huang S, Sasaki A, Yamaji N, Okada H, Mitani-Ueno N, Ma J F. 2020. The ZIP transporter family member OsZIP9 contributes to root zinc uptake in rice under zinc-limited conditions. Plant Physiol, 183(3): 1224-1234.
PMID |
| [21] |
Huizar M I, Arena R, Laddu D R. 2021. The global food syndemic: The impact of food insecurity, malnutrition and obesity on the healthspan amid the COVID-19 pandemic. Prog Cardiovasc Dis, 64: 105-107.
PMID |
| [22] | Impa S M, Morete M J, Ismail A M, Schulin R, Johnson-Beebout S E. 2013. Zn uptake, translocation and grain Zn loading in rice (Oryza sativa L.) genotypes selected for Zn deficiency tolerance and high grain Zn. J Exp Bot, 64(10): 2739-2751. |
| [23] | Ishimaru Y, Bashir K, Nishizawa N K. 2011. Zn uptake and translocation in rice plants. Rice, 4(1): 21-27. |
| [24] |
Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa N K. 2005. OsZIP4, a novel zinc-regulated zinc transporter in rice. J Exp Bot, 56: 3207-3214.
PMID |
| [25] |
Kitomi Y, Kitano H, Inukai Y. 2011. Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal Behav, 6(9): 1270-1278.
PMID |
| [26] | Kumar Sarmah C, Samarasinghe S. 2010. Microarray data integration: Frameworks and a list of underlying issues. Curr Bioinform, 5(4): 280-289. |
| [27] |
Langfelder P, Horvath S. 2008. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9: 559.
PMID |
| [28] | Lee S, Chiecko J C, Kim S A, Walker E L, Lee Y, Guerinot M L, An G. 2009. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol, 150(2): 786-800. |
| [29] | Lee S, Kim S A, Lee J, Guerinot M L, An G. 2010. Zinc deficiency- inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells, 29(6): 551-558. |
| [30] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25(16): 2078-2079.
PMID |
| [31] |
Liao Y, Smyth G K, Shi W. 2014. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7): 923-930.
PMID |
| [32] | Lu X, Liu S, Zhi S, Chen J, Ye G. 2021. Comparative transcriptome profile analysis of rice varieties with different tolerance to zinc deficiency. Plant Biol, 23(2): 375-390. |
| [33] |
Luo J, Schumacher M, Scherer A, Sanoudou D, Megherbi D, Davison T, Shi T, Tong W, Shi L, Hong H, Zhao C, Elloumi F, Shi W, Thomas R, Lin S, Tillinghast G, Liu G, Zhou Y, Herman D, Li Y, Deng Y, Fang H, Bushel P, Woods M, Zhang J. 2010. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J, 10(4): 278-291.
PMID |
| [34] | Lv Y M, Xu L, Dossa K, Zhou K, Zhu M D, Xie H J, Tang S J, Yu Y Y, Guo X Y, Zhou B. 2019. Identification of putative drought- responsive genes in rice using gene co-expression analysis. Bioinformation, 15(7): 480-489. |
| [35] | Maurya S, Vishwakarma A K, Dubey M, Shrivastava P, Shrivastava R, Chandel G. 2018. Developing gene-tagged molecular marker for functional analysis of OsZIP10 metal transporter gene in rice. Indian J Genet Plant Breed, 78(2): 180. |
| [36] |
Meuwissen T H, Hayes B J, Goddard M E. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 157(4): 1819-1829.
PMID |
| [37] |
Nanda A K, Wissuwa M. 2016. Rapid crown root development confers tolerance to zinc deficiency in rice. Front Plant Sci, 7: 428.
PMID |
| [38] |
Nanda A K, Pujol V, Wissuwa M. 2017. Patterns of stress response and tolerance based on transcriptome profiling of rice crown tissue under zinc deficiency. J Exp Bot, 68(7): 1715-1729.
PMID |
| [39] |
Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa N K. 2011. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J Biol Chem, 286(7): 5446-5454.
PMID |
| [40] |
Ogo Y, Itai R N, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa N K. 2006. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot, 57(11): 2867-2878.
PMID |
| [41] |
Olsen L I, Palmgren M G. 2014. Many rivers to cross: The journey of zinc from soil to seed. Front Plant Sci, 5: 30.
PMID |
| [42] |
Pertea M, Kim D, Pertea G M, Leek J T, Salzberg S L. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc, 11(9): 1650-1667.
PMID |
| [43] | Ptashnyk M, Roose T, Jones D L, Kirk G J D. 2011. Enhanced zinc uptake by rice through phytosiderophore secretion: A modelling study. Plant Cell Environ, 34(12): 2038-2046. |
| [44] |
Qi Y H, Wang S K, Shen C J, Zhang S N, Chen Y, Xu Y X, Liu Y, Wu Y R, Jiang D A. 2012. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol, 193(1): 109-120.
PMID |
| [45] | Ramírez-González R H, Borrill P, Lang D, Harrington S A, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, Khedikar Y, Robinson S J, Cory A T, Florio T, Concia L, Juery C, Schoonbeek H, Steuernagel B, Xiang D, Ridout C J, Chalhoub B, Mayer K F X, Benhamed M, Latrasse D, Bendahmane A, Consortium I W G S, Wulff B B H, Appels R, Tiwari V, Datla R, Choulet F, Pozniak C J, Provart N J, Sharpe A G, Paux E, Spannagl M, Bräutigam A, Uauy C. 2018. The transcriptional landscape of polyploid wheat. Science, 361: eaar6089. |
| [46] | Rani Debi B, Taketa S, Ichii M. 2005. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J Plant Physiol, 162(5): 507-515. |
| [47] | Ritchie M E, Phipson B, Wu D, Hu Y F, Law C W, Shi W, Smyth G K. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43(7): e47. |
| [48] |
Schmittgen T D, Livak K J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc, 3(6): 1101-1108.
PMID |
| [49] |
Sinclair S A, Krämer U. 2012. The zinc homeostasis network of land plants. Biochim Biophys Acta, 1823(9): 1553-1567.
PMID |
| [50] |
Singh S P, Gruissem W, Bhullar N K. 2017. Single genetic locus improvement of iron, zinc and β-carotene content in rice grains. Sci Rep, 7(1): 6883.
PMID |
| [51] |
Spindel J E, Begum H, Akdemir D, Collard B, Redoña E, Jannink J L, McCouch S. 2016. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity, 116(4): 395-408.
PMID |
| [52] |
Suzuki M, Tsukamoto T, Inoue H, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa N K. 2008. Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol Biol, 66(6): 609-617.
PMID |
| [53] |
Swamy B P M, Rahman M A, Inabangan-Asilo M A, Amparado A, Manito C, Chadha-Mohanty P, Reinke R, Slamet-Loedin I H. 2016. Advances in breeding for high grain zinc in rice. Rice, 9(1): 49.
PMID |
| [54] |
Tan M P, Cheng D, Yang Y N, Zhang G Q, Qin M J, Chen J, Chen Y H, Jiang M Y. 2017. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol, 17(1): 194.
PMID |
| [55] |
Tian D G, Chen Z J, Lin Y, Chen Z Q, Bui K T, Wang Z H, Wang F. 2020. Weighted gene co-expression network coupled with a critical-time-point analysis during pathogenesis for predicting the molecular mechanism underlying blast resistance in rice. Rice, 13(1): 81.
PMID |
| [56] |
van der Straeten D, Bhullar N K, de Steur H, Gruissem W, MacKenzie D, Pfeiffer W, Qaim M, Slamet-Loedin I, Strobbe S, Tohme J, Trijatmiko K R, Vanderschuren H, van Montagu M, Zhang C Y, Bouis H. 2020. Multiplying the efficiency and impact of biofortification through metabolic engineering. Nat Commun, 11(1): 5203.
PMID |
| [57] | Wang A J, Shu X Y, Niu X Y, Zhao W J, Ai P, Li P, Zheng A P. 2018. Comparison of gene co-networks analysis provide a systems view of rice (Oryza sativa L.) response to Tilletia horrida infection. PLoS One, 13(10): e0202309. |
| [58] | Wang Y, Frei M, Wissuwa M. 2008. An agar nutrient solution technique as a screening tool for tolerance to zinc deficiency and iron toxicity in rice. Soil Sci Plant Nutr, 54(5): 744-750. |
| [59] | White P J, Broadley M R. 2009. Biofortification of crops with seven mineral elements often lacking in human diets: Iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol, 182(1): 49-84. |
| [60] | Wissuwa M, Ismail A M, Graham R D. 2008. Rice grain zinc concentrations as affected by genotype, native soil-zinc availability, and zinc fertilization. Plant Soil, 306: 37-48. |
| [61] | Xu Y, Ma K X, Zhao Y, Wang X, Zhou K, Yu G N, Li C, Li P C, Yang Z F, Xu C W, Xu S Z. 2021. Genomic selection: A breakthrough technology in rice breeding. Crop J, 9(3): 669-677. |
| [62] | Yang M, Li Y T, Liu Z H, Tian J J, Liang L M, Qiu Y, Wang G Y, Du Q Q, Cheng D, Cai H M, Shi L, Xu F S, Lian X M. 2020. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J, 103(5): 1695-1709. |
| [63] |
Yang Z, Wu Y R, Li Y, Ling H Q, Chu C C. 2009. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol, 70: 219-229.
PMID |
| [64] |
Yao W, Li G W, Yu Y M, Ouyang Y D. 2018. funRiceGenes dataset for comprehensive understanding and application of rice functional genes. GigaScience, 7(1): 1-9.
PMID |
| [65] | Yi X, Du Z, Su Z. 2013. PlantGSEA: A gene set enrichment analysis toolkit for plant community. Nucleic Acids Res, 41: W98-W103. |
| [66] |
Yu G C, Wang L G, Han Y Y, He Q Y. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16(5): 284-287.
PMID |
| [67] | Zeng H Q, Wang G P, Hu X Y, Wang H Z, Du L Q, Zhu Y Y. 2014. Role of microRNAs in plant responses to nutrient stress. Plant Soil, 374: 1005-1021. |
| [68] | Zeng H Q, Zhang X, Ding M, Zhang X J, Zhu Y Y. 2019a. Transcriptome profiles of soybean leaves and roots in response to zinc deficiency. Physiol Plant, 167(3): 330-351. |
| [69] | Zeng H Q, Zhang X, Ding M, Zhu Y Y. 2019b. Integrated analyses of miRNAome and transcriptome reveal zinc deficiency responses in rice seedlings. BMC Plant Biol, 19(1): 585. |
| [70] | Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol, 4: Article17. |
| [71] | Zhang Y Q, Parmigiani G, Johnson W E. 2020. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom Bioinform, 2(3): lqaa078. |
| [72] |
Zheng Y, Jiao C, Sun H H, Rosli H G, Pombo M A, Zhang P F, Banf M, Dai X B, Martin G B, Giovannoni J J, Zhao P X, Rhee S Y, Fei Z J. 2016. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant, 9(12): 1667-1670.
PMID |
| [73] |
Zhou W, Koudijs K K M, Böhringer S. 2019. Influence of batch effect correction methods on drug induced differential gene expression profiles. BMC Bioinformatics, 20(1): 437.
PMID |
| [74] | Zhu M D, Xie H J, Wei X J, Dossa K, Yu Y Y, Hui S Z, Tang G H, Zeng X S, Yu Y H, Hu P S, Wang J L. 2019. WGCNA analysis of salt-responsive core transcriptome identifies novel hub genes in rice. Genes, 10(9): 719. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||