
Rice Science ›› 2023, Vol. 30 ›› Issue (3): 222-234.DOI: 10.1016/j.rsci.2023.03.006
• Research Paper • Previous Articles Next Articles
Jiang Hongzhen1,#, Wang Yamei1,#, Lai Liuru1, Liu Xintong1, Miao Changjian1, Liu Ruifang2, Li Xiaoyun1, Tan Jinfang1, Gao Zhenyu3( ), Chen Jingguang1(
), Chen Jingguang1( )
)
Received:2022-10-17
Accepted:2023-01-14
Online:2023-05-28
Published:2023-02-21
Contact:
Chen Jingguang (chenjg28@mail.sysu.edu.cn); Gao Zhenyu (gaozhenyu@caas.cn)
About author:First author contact:#These authors contributed equally to this work
Jiang Hongzhen, Wang Yamei, Lai Liuru, Liu Xintong, Miao Changjian, Liu Ruifang, Li Xiaoyun, Tan Jinfang, Gao Zhenyu, Chen Jingguang. OsAMT1.1 Expression by Nitrate-Inducible Promoter of OsNAR2.1 Increases Nitrogen Use Efficiency and Rice Yield[J]. Rice Science, 2023, 30(3): 222-234.
Add to citation manager EndNote|Ris|BibTeX
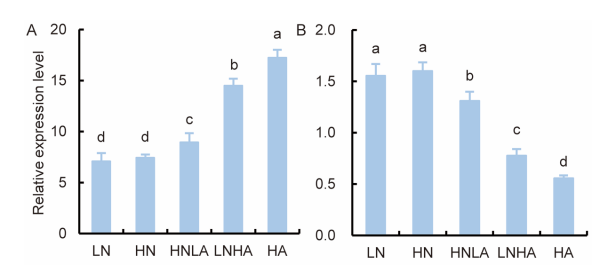
Fig. 1. Effects of nitrogen levels on transcriptional expression of OsAMT1.1 and OsNAR2.1 in rice roots. A, Expression of OsAMT1.1. B, Expression of OsNAR2.1.Nipponbare seedlings were cultured in the IRRI solution containing 1.0 mmol/L NH4NO3 for three weeks and shifted to different N supply levels for one additional week. LN, 0.5 mmol/L NO3-; HN, 2.5 mmol/L NO3-; HNLA, 2.0 mmol/L NO3- + 0.5 mmol/L NH4+; LNHA, 0.5 mmol/L NO3- + 2.0 mmol/L NH4+; HA, 2.5 mmol/L NH4+. Actin1 gene of rice was used as an internal control. Data are Mean ± SE (n = 3). Different lowercase letters above the bars indicate significant differences at the 0.05 level.
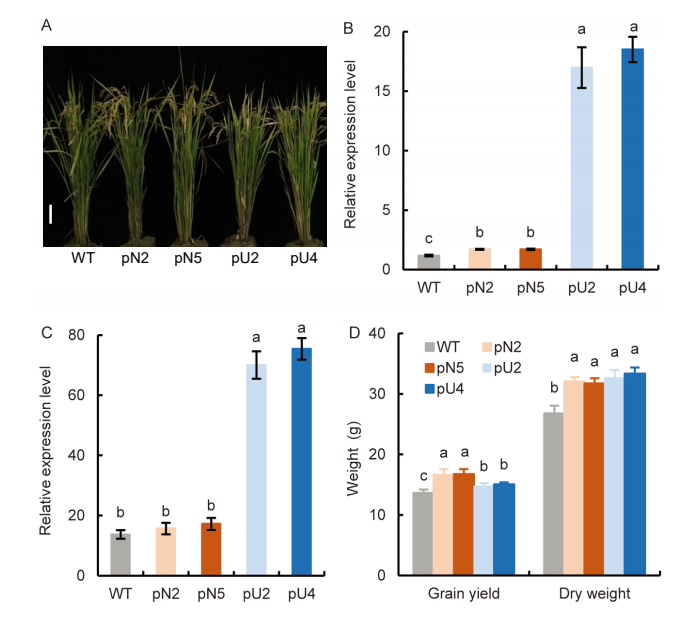
Fig. 2. Characterization of pOsNAR2.1: OsAMT1.1 and pUbi:OsAMT1.1 transgenic lines (T3 generation). A, Gross morphology of wild type (WT), pOsNAR2.1:OsAMT1.1 (pN2 and pN5) and pUbi:OsAMT1.1 (pU2 and pU4) transgenic lines. Scale bar, 10 cm. B and C, Endogenous OsAMT1.1 expression in root (B) and leaf blade I (C). Actin1 gene of rice was used as an internal control. Data are Mean ± SE (n = 3).D, Grain yield and dry weight per plant. Mean dry weight values referred to all the biomass aboveground, including grain yield. Data are Mean ± SE (n = 4). Different lowercase letters above the bars for the same trait indicate significant differences at the 0.05 level.
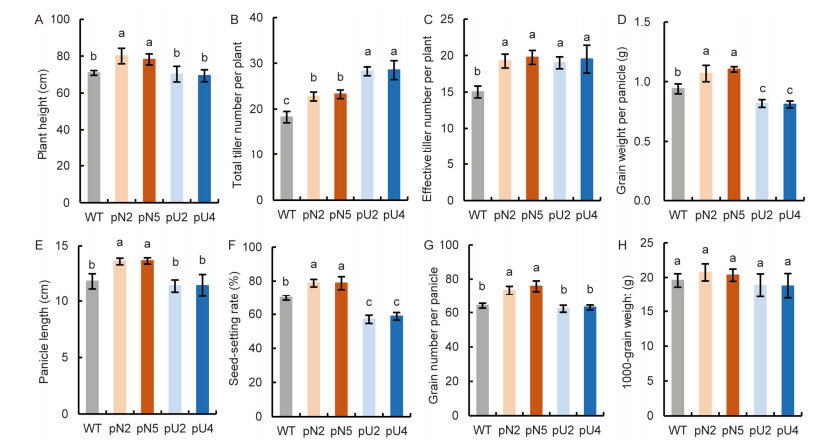
Fig. 3. Comparison of agronomic traits in transgenic lines (T3 generation). A, Plant height. B, Total tiller number per plant. C, Effective tiller number per plant. D, Grain weight per panicle. E, Panicle length. F, Seed-setting rate. G, Grain number per panicle. H, 1000-grain weight. WT, Wild type; pN2 and pN5 are pOsNAR2.1:OsAMT1.1 transgenic lines while pU2 and pU4 are pUbi:OsAMT1.1 transgenic lines.Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at the 0.05 level.

Fig. 4. Biomass and nitrogen (N) content in different parts of transgenic lines at flowering and maturity stages in fields. A and B, WT and T4 generation transgenic plants in the field. C-E, Dry weight (C), total N concentration (D) and total N content per plant (E) in different parts of transgenic lines and WT at the flowering stage. F-H, Dry weight (F), total N concentration (G) and total N content (H) in different parts of transgenic lines and WT at the maturity stage. WT, Wild type; pN2 and pN5 are pOsNAR2.1:OsAMT1.1 transgenic lines while pU2 and pU4 are pUbi:OsAMT1.1 transgenic lines.Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at the 0.05 level.
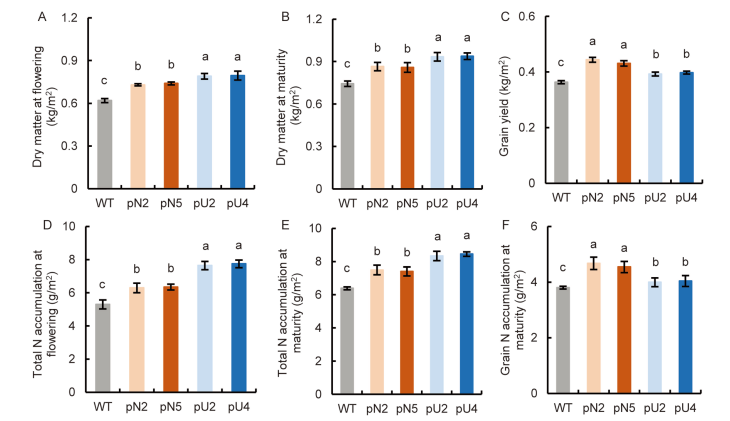
Fig. 5. Biomass and nitrogen (N) accumulation of transgenic lines (T4 generation) in fields. A, Dry matter at the flowering stage. B, Dry matter at the maturity stage. C, Grain yield. D, Total N accumulation at the flowering stage. E, Total N accumulation at the maturity stage. F, Grain N accumulation at the maturity stage. WT, Wild type; pN2 and pN5 are pOsNAR2.1:OsAMT1.1 transgenic lines while pU2 and pU4 are pUbi:OsAMT1.1 transgenic lines.Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at the 0.05 level.
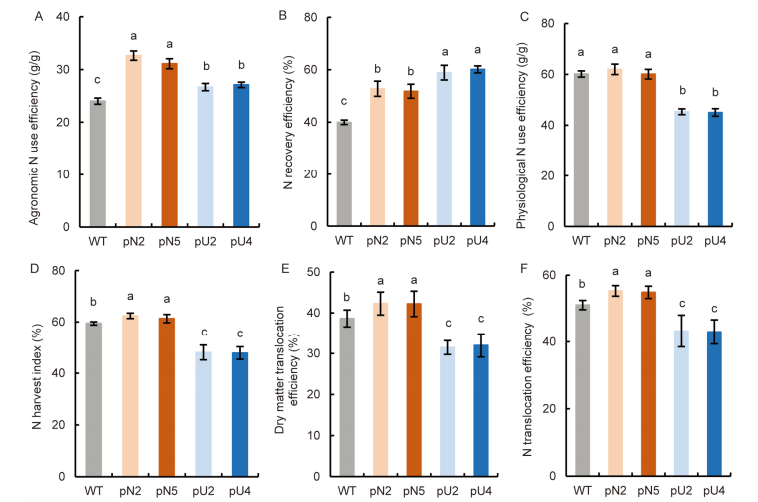
Fig. 6. Comparison of nitrogen (N) use efficiency between WT and transgenic lines (T4 generation). A, Agronomic N use efficiency. B, N recovery efficiency. C, Physiological N use efficiency. D, N harvest index. E, Dry matter translocation efficiency. F, N translocation efficiency. WT, Wild type; pN2 and pN5 are pOsNAR2.1:OsAMT1.1 transgenic lines while pU2 and pU4 are pUbi:OsAMT1.1 transgenic lines.Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at the 0.05 level.
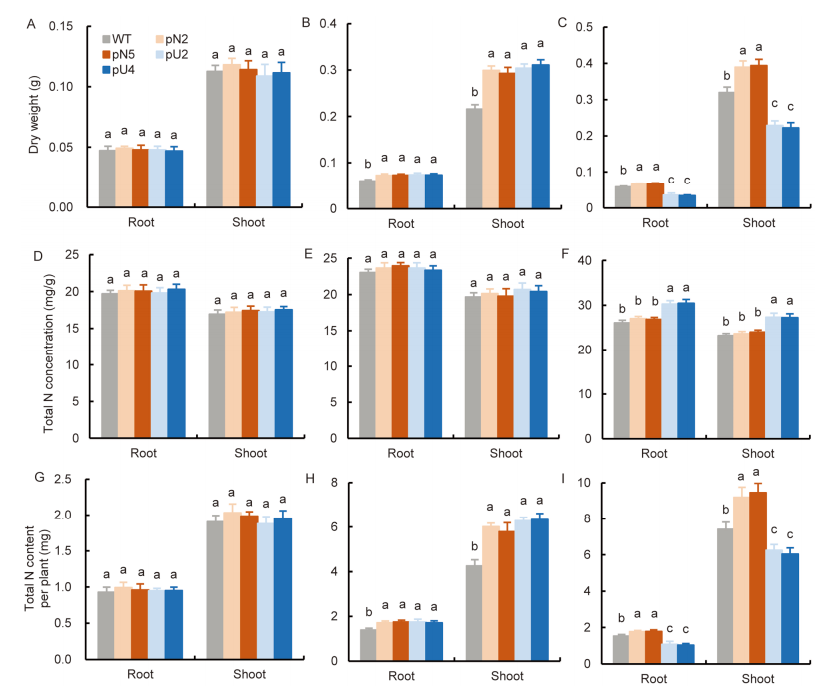
Fig. 7. Comparison of growth and total nitrogen (N) content of transgenic lines at different N supply levels. A-C, Dry weights of seedlings treated with 0.5 mmol/L NO3- (A), 2.0 mmol/L NO3- + 0.5 mmol/L NH4+ (B), and 0.5 mmol/L NO3- + 2.0 mmol/L NH4+ (C).D-F, Total N concentration in roots and shoots in the pN lines, pU lines and WT under 0.5 mmol/L NO3- (D), 2.0 mmol/L NO3- + 0.5 mmol/L NH4+ (E), and 0.5 mmol/L NO3- + 2.0 mmol/L NH4+ (F). G-I, Total N content of roots and shoots, under 0.5 mmol/L NO3- (G), 2.0 mmol/L NO3- + 0.5 mmol/L NH4+ (H), and 0.5 mmol/L NO3- + 2.0 mmol/L NH4+ (I). WT, Wild type; pN2 and pN5 are pOsNAR2.1:OsAMT1.1 transgenic lines while pU2 and pU4 are pUbi:OsAMT1.1 transgenic lines.WT and transgenic rice seedlings were cultured in the IRRI solution containing 1.0 mmol/L NO3- for one week and then in different forms of N for two additional weeks.Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at the 0.05 level.
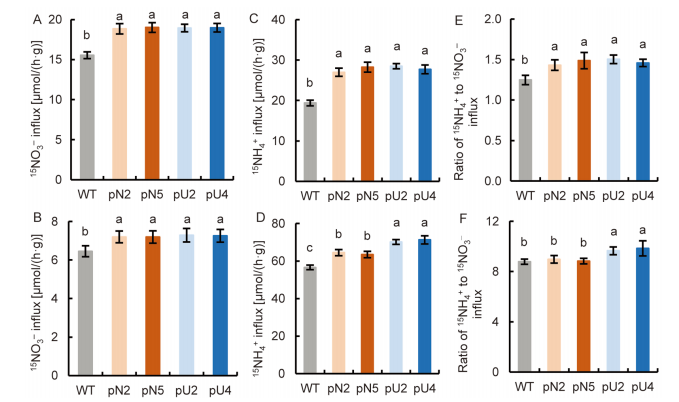
Fig. 8. NH4+ and NO3- influx rates in transgenic lines using 15N enriched sources. A-C, 15NO3- influx rate (A), 15NH4+ influx rate (B) and ratio of 15NH4+ to 15NO3- influx (C) at 2.0 mmol/L 15NO3- + 0.5 mmol/L NH4+.D-F, 15NO3- influx rate (D), 15NH4+ influx rate (E) and ratio of 15NH4+ to 15NO3- influx (F) at 0.5 mmol/L 15NO3- + 2.0 mmol/L NH4+. Wild type (WT) and transgenic seedlings were grown in 1.0 mmol/L NO3- for three weeks and N starved for 3 d. Data are Mean ± SE (n = 4). Different lowercase letters above the bars indicate significant differences at P ≤ 0.05.
| [1] | Briones A M, Okabe S, Umemiya Y, Ramsing N B, Reichardt W, Okuyama H. 2002. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl Environ Microbiol, 68(6): 3067-3075. |
| [2] | Britto D T, Kronzucker H J. 2002. NH4+ toxicity in higher plants: A critical review. J Plant Physiol, 159(6): 567-584. |
| [3] | Chen J G, Zhang Y, Tan Y W, Zhang M, Zhu L L, Xu G H, Fan X R. 2016. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol J, 14(8): 1705-1715. |
| [4] | Chen J G, Fan X R, Qian K Y, Zhang Y, Song M Q, Liu Y, Xu G H, Fan X R,. 2017. pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol J, 15(10): 1273-1283. |
| [5] | Chen J G, Liu X Q, Liu S H, Fan X R, Zhao L M, Song M Q, Fan X R, Xu G H. 2020. Co-overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front Plant Sci, 11: 1245. |
| [6] | Coleto I, Bejarano I, Marín-Peña A J, Medina J, Rioja C, Burow M, Marino D. 2021. Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol, 229(2): 1021-1035. |
| [7] | Crawford N M, Forde B G. 2002. Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book, 1: e0011. |
| [8] |
Esteban R, Ariz I, Cruz C, Moran J F. 2016. Review: Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci, 248: 92-101.
PMID |
| [9] |
Fan X R, Tang Z, Tan Y W, Zhang Y, Luo B B, Yang M, Lian X M, Shen Q R, Miller A J, Xu G H. 2016. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA, 113(26): 7118-7123.
PMID |
| [10] | Fang G, Yang J, Sun T, Wang X X, Li Y S. 2021. Evidence that synergism between potassium and nitrate enhances the alleviation of ammonium toxicity in rice seedling roots. PLoS One, 16(9): e0248796. |
| [11] |
Feng H M, Yan M, Fan X R, Li B Z, Shen Q R, Miller A J, Xu G H. 2011. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot, 62(7): 2319-2332.
PMID |
| [12] | Gao Z Y, Wang Y F, Chen G, Zhang A P, Yang S L, Shang L G, Wang D Y, Ruan B P, Liu C L, Jiang H Z, Dong G J, Zhu L, Hu J, Zhang G H, Zeng D L, Guo L B, Xu G H, Teng S, Harberd N P, Qian Q. 2019. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat Commun, 10(1): 5207. |
| [13] | Garnett T, Conn V, Kaiser B N. 2009. Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ, 32(9): 1272-1283. |
| [14] |
Good A G, Shrawat A K, Muench D G. 2004. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci, 9(12): 597-605.
PMID |
| [15] | Hachiya T, Watanabe C K, Fujimoto M, Ishikawa T, Takahara K, Kawai-Yamada M, Uchimiya H, Uesono Y, Terashima I, Noguchi K. 2012. Nitrate addition alleviates ammonium toxicity without lessening ammonium accumulation, organic acid depletion and inorganic cation depletion in Arabidopsis thaliana shoots. Plant Cell Physiol, 53(3): 577-591. |
| [16] |
Hachiya T, Sakakibara H. 2017. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J Exp Bot, 68(10): 2501-2512.
PMID |
| [17] | Hirel B, Tétu T, Lea P J, Dubois F. 2011. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability, 3(9): 1452-1485. |
| [18] | Hoque M S, Masle J, Udvardi M K, Ryan P R, Upadhyaya N M. 2006. Over-expression of the rice OsAMT1-1 gene increases ammonium uptake and content, but impairs growth and development of plants under high ammonium nutrition. Funct Plant Biol, 33(2): 153-163. |
| [19] | Hou M M, Yu M, Li Z Q, Ai Z Y, Chen J G. 2021. Molecular regulatory networks for improving nitrogen use efficiency in rice. Int J Mol Sci, 22(16): 9040. |
| [20] | Hu B, Wang W, Ou S J, Tang J Y, Li H, Che R H, Zhang Z H, Chai X Y, Wang H R, Wang Y Q, Liang C Z, Liu L C, Piao Z Z, Deng Q Y, Deng K, Xu C, Liang Y, Zhang L H, Li L G, Chu C C. 2015. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet, 47(7): 834-838. |
| [21] | Jia L T, Xie Y M, Wang Z, Luo L, Zhang C, Pélissier P M, Parizot B, Qi W C, Zhang J, Hu Z B, Motte H, Luo L, Xu G H, Beeckman T, Xuan W. 2020. Rice plants respond to ammonium stress by adopting a helical root growth pattern. Plant J, 104(4): 1023-1037. |
| [22] | Khademi S, O'Connell J III, Remis J, Robles-Colmenares Y, Miercke L J W, Stroud R M. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. Science, 305: 1587-1594. |
| [23] | Kirk G D, Kronzucker H J. 2005. The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: A modelling study. Ann Bot, 96(4): 639-646. |
| [24] |
Konishi N, Ma J F. 2021. Three polarly localized ammonium transporter 1 members are cooperatively responsible for ammonium uptake in rice under low ammonium condition. New Phytol, 232(4): 1778-1792.
PMID |
| [25] |
Kronzucker H J, Siddiqi M Y, Glass A D, Kirk G J. 1999. Nitrate-ammonium synergism in rice: A subcellular flux analysis. Plant Physiol, 119(3): 1041-1046.
PMID |
| [26] | Kumar A, Kaiser B N, Siddiqi M Y, Glass A D M. 2006. Functional characterisation of OsAMT1.1 overexpression lines of rice, Oryza sativa. Funct Plant Biol, 33(4): 339-346. |
| [27] | Lee S, Marmagne A, Park J, Fabien C, Yim Y, Kim S J, Kim T H, Lim P O, Masclaux-Daubresse C, Nam H G. 2020. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J, 103(1): 7-20. |
| [28] | Li B H, Li G J, Kronzucker H J, Baluška F, Shi W M. 2014. Ammonium stress in Arabidopsis: Signaling, genetic loci, and physiological targets. Trends Plant Sci, 19(2): 107-114. |
| [29] | Li L, Zhang Z, Tian H, Ashraf U, Hamoud Y A, Alaa A A, Tang X R, Duan M Y, Wang Z M, Pan S G. 2022. Nitrogen deep placement combined with straw mulch cultivation enhances physiological traits, grain yield and nitrogen use efficiency in mechanical pot-seedling transplanting rice. Rice Sci, 29(1): 89-100. |
| [30] | Li S, Tian Y H, Wu K, Ye Y F, Yu J P, Zhang J Q, Liu Q, Hu M Y, Li H, Tong Y P, Harberd N P, Fu X D. 2018. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature, 560: 595-600. |
| [31] | Li Y L, Fan X R, Shen Q R. 2008. The relationship between rhizosphere nitrification and nitrogen use efficiency in rice plants. Plant Cell Environ, 31(1): 73-85. |
| [32] | Liu X Q, Feng H M, Huang D M, Song M Q, Fan X R, Xu G H. 2015. Two short sequences in OsNAR2.1 promoter are necessary for fully activating the nitrate induced gene expression in rice roots. Sci Rep, 5(1): 11950. |
| [33] | Luo L, Zhu M, Jia L T, Xie Y M, Wang Z N, Xuan W. 2022. Ammonium transporters cooperatively regulate rice crown root formation responding to ammonium nitrogen. J Exp Bot, 73(11): 3671-3685. |
| [34] |
Miller A J, Fan X R, Orsel M, Smith S J, Wells D M. 2007. Nitrate transport and signalling. J Exp Bot, 58(9): 2297-2306.
PMID |
| [35] | Pastor V, Gamir J, Camañes G, Cerezo M, Sánchez-Bel P, Flors V. 2014. Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front Plant Sci, 5: 231. |
| [36] |
Ranathunge K, El-Kereamy A, Gidda S, Bi Y M, Rothstein S J. 2014. AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J Exp Bot, 65(4): 965-979.
PMID |
| [37] | Scheible W R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi M K, Stitt M. 2004. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol, 136(1): 2483-2499. |
| [38] | Sonoda Y, Ikeda A, Saiki S, von Wirén N, Yamaya T, Yamaguchi J. 2003. Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol, 44(7): 726-734. |
| [39] | Sun L, Di D W, Li G J, Kronzucker H J, Shi W M. 2017. Spatio-temporal dynamics in global rice gene expression (Oryza sativa L.) in response to high ammonium stress. J Plant Physiol, 212: 94-104. |
| [40] |
Swarbreck S M, Wang M, Wang Y, Kindred D, Sylvester-Bradley R, Shi W M, Varinderpal-Singh, Bentley A R, Griffiths H. 2019. A roadmap for lowering crop nitrogen requirement. Trends Plant Sci, 24(10): 892-904.
PMID |
| [41] |
Tang W J, Ye J, Yao X M, Zhao P Z, Xuan W, Tian Y L, Zhang Y Y, Xu S, An H Z, Chen G M, Yu J, Wu W, Ge Y W, Liu X L, Li J, Zhang H Z, Zhao Y Q, Yang B, Jiang X Z, Peng C, Zhou C, Terzaghi W, Wang C M, Wan J M. 2019. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat Commun, 10(1): 5279.
PMID |
| [42] | Wang W, Hu B, Yuan D Y, Liu Y Q, Che R H, Hu Y C, Ou S J, Liu Y X, Zhang Z H, Wang H R, Li H, Jiang Z M, Zhang Z L, Gao X K, Qiu Y H, Meng X B, Liu Y X, Bai Y, Liang Y, Wang Y Q, Zhang L H, Li L G, Sodmergen, Jing H C, Li J Y, Chu C C. 2018. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell, 30(3): 638-651. |
| [43] | Wu J, Zhang Z S, Xia J Q, Alfatih A, Song Y, Huang Y J, Wan G Y, Sun L Q, Tang H, Liu Y, Wang S M, Zhu Q S, Qin P, Wang Y P, Li S G, Mao C Z, Zhang G Q, Chu C C, Yu L H, Xiang C B. 2021. Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol J, 19(3): 448-461. |
| [44] | Wu K, Wang S S, Song W Z, Zhang J Q, Wang Y, Liu Q, Yu J P, Ye Y F, Li S, Chen J F, Zhao Y, Wang J, Wu X K, Wang M Y, Zhang Y J, Liu B M, Wu Y J, Harberd N P, Fu X D. 2020. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science, 367: eaaz2046. |
| [45] |
Wu X X, Yuan D P, Chen H, Kumar V, Kang S M, Jia B L, Xuan Y H. 2022. Ammonium transporter 1 increases rice resistance to sheath blight by promoting nitrogen assimilation and ethylene signalling. Plant Biotechnol J, 20(6): 1085-1097.
PMID |
| [46] | Xu N, Yu B, Chen R R, Li S T, Zhang G C, Huang J L. 2020. OsNAR2.2 plays a vital role in the root growth and development by promoting nitrate uptake and signaling in rice. Plant Physiol Biochem, 149: 159-169. |
| [47] | Xu Y J, Tang S P, Jian C Q, Cai W L, Zhang W Y, Wang Z Q, Yang J C. 2022. Roles of polyamines and ethylene in grain filling, grain weight and quality of rice. Chin J Rice Sci, 36(4): 327-335. (in Chinese with English abstract) |
| [48] |
Xuan W, Beeckman T, Xu G H. 2017. Plant nitrogen nutrition: Sensing and signaling. Curr Opin Plant Biol, 39: 57-65.
PMID |
| [49] | Yan M, Fan X R, Feng H M, Miller A J, Shen Q R, Xu G H. 2011. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ, 34(8): 1360-1372. |
| [50] | Yu J, Xuan W, Tian Y L, Fan L, Sun J, Tang W J, Chen G M, Wang B X, Liu Y, Wu W, Liu X L, Jiang X Z, Zhou C, Dai Z Y, Xu D Y, Wang C M, Wan J M. 2021. Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice. Plant Biotechnol J, 19(1): 167-176. |
| [51] | Zhang C, Rees R M, Ju X T. 2021. Cropping system design can improve nitrogen use efficiency in intensively managed agriculture. Environ Pollut, 280: 116967. |
| [52] | Zhang Z S, Xia J Q, Alfatih A, Song Y, Huang Y J, Sun L Q, Wan G Y, Wang S M, Wang Y P, Hu B H, Zhang G H, Qin P, Li S G, Yu L H, Wu J, Xiang C B. 2022. Rice NIN-LIKE PROTEIN 3 modulates nitrogen use efficiency and grain yield under nitrate-sufficient conditions. Plant Cell Environ, 45(5): 1520-1536. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||