
Rice Science ›› 2025, Vol. 32 ›› Issue (2): 243-258.DOI: 10.1016/j.rsci.2024.11.002
• Research Papers • Previous Articles Next Articles
He Zhenrui1,#, Zhao Wenhua1,#, Cheng Baoping2, Yang Mei1, Yang Yingqing3( ), Zhu Yiming1(
), Zhu Yiming1( ), Zhou Erxun1(
), Zhou Erxun1( )
)
Received:2024-08-21
Accepted:2024-11-23
Online:2025-03-28
Published:2025-04-14
Contact:
Zhou Erxun (exzhou@scau.edu.cn); Zhu Yiming (zhu_yiming1992@scau.edu.cn); Yang Yingqing (yyq8295@163.com)
About author: These authors contributed equally to this work
He Zhenrui, Zhao Wenhua, Cheng Baoping, Yang Mei, Yang Yingqing, Zhu Yiming, Zhou Erxun. Molecular and Biological Characterization of Novel Mitovirus Infecting Phytopathogenic Fungus Ustilaginoidea virens[J]. Rice Science, 2025, 32(2): 243-258.
Add to citation manager EndNote|Ris|BibTeX
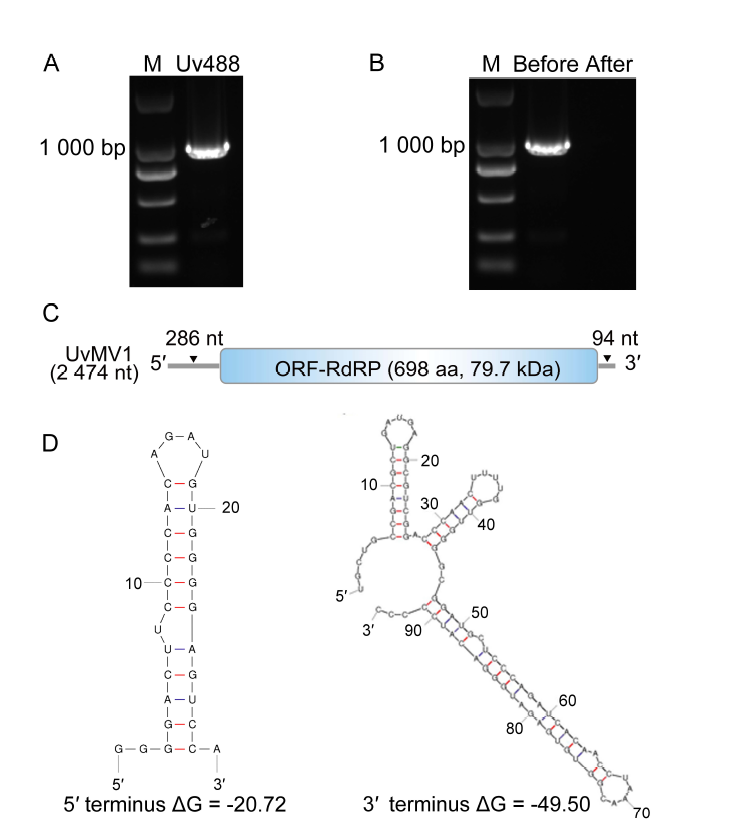
Fig. 1. Characterization of Ustilaginoidea virens mitovirus 1 (UvMV1). A, RT-PCR was conducted to identify potential mycoviruses in U. virens strain Uv488. M, DNA marker (DL2000 bp). B, RT-PCR was conducted on the viral genome using mycovirus- specific primers both before and after treatments with S1 nuclease for the digestion of ssDNA or ssRNA. M, DNA marker (DL2000 bp).C, Schematic illustrates the genome organization of UvMV1, with boxes representing putative open reading frame (ORF) encoding an RNA-dependent RNA polymerase (RdRp), and gray lines indicate untranslated regions. D, Predicted secondary structure of the 5ʹ and 3ʹ terminal sequences of UvMV1.
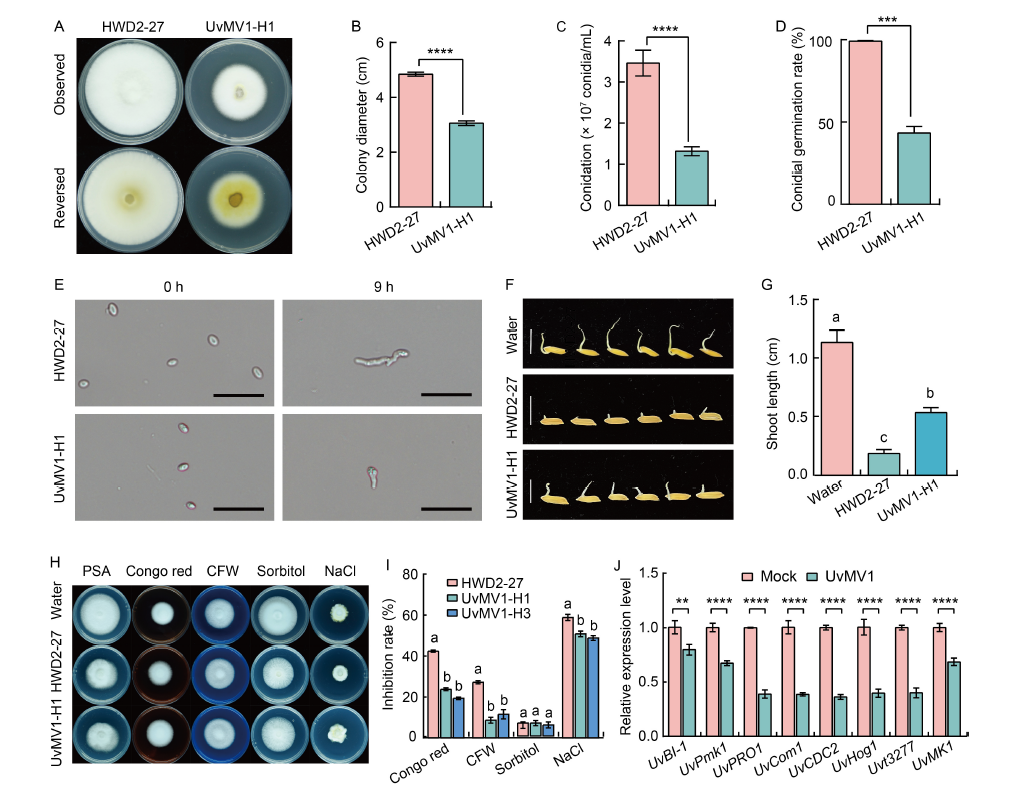
Fig. 2. Effect of UvMV1 on biological characteristics of Ustilaginoidea virens. A, Colony morphology of the UvMV1-free strain HWD2-27 compared with UvMV1-infected strain UvMV1-H1 grown on potato sucrose agar (PSA) plates at 28 ºC for 14 d. B, Comparison of colony diameter of strains HWD2-27 and UvMV1-H1 grown on PSA plates for 14 d. C, Comparison of the number of conidia produced by strains HWD2-27 and UvMV1-H1 when cultured on potato sucrose (PS) liquid medium under shaking conditions for 7 d. D, Conidial germination rates of strains HWD2-27 and UvMV1-H1 at 9 h after incubation.E, Spores of strains HWD2-27 and UvMV1-H1 were examined using microscopy at both 0 and 9 h after incubation. Scale bars, 20 μm. F, Toxicity assessment of culture filtrates from 7-day-old strains HWD2-27 and UvMV1-H1 to evaluate their effects on rice seed germination. Scale bars, 1 cm. G, Shoot lengths of rice seeds following a 5-day treatment with culture filtrates derived from strains HWD2-27 and UvMV1-H1. H and I, Colony morphologies (H) and growth inhibition rates (I) of UvMV1-free strain HWD2-27 and UvMV1-infected strains UvMV1-H1 and UvMV1-H3 after culturing on PSA medium plates containing various environmental stress factors [120 μg/mL Congo red, 120 μg/mL Calcofluor white (CFW), 0.5 mol/L sorbitol, or 0.3 mol/L NaCl] for 14 d. J, qRT-PCR analysis of the transcript levels of UvBI-1, UvPmk1, UvPRO1, UvCom1, UvCDC2, UvHog1, Uvt3277, and UvMK1 in mock- and UvMV1-infected U. virens. The mock- and UvMV1-infected strains were cultured in PS liquid medium for 5 d with shaking in the dark, after which total RNA was extracted for qRT-PCR analyses. The housekeeping gene β-tubulin serves as the internal control. Data are Mean ± SD (n = 3) in B‒D, G, I, and J. Student’s t-test was used for analyses: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Lowercase letters above the bars indicate significant differences among the treatments.
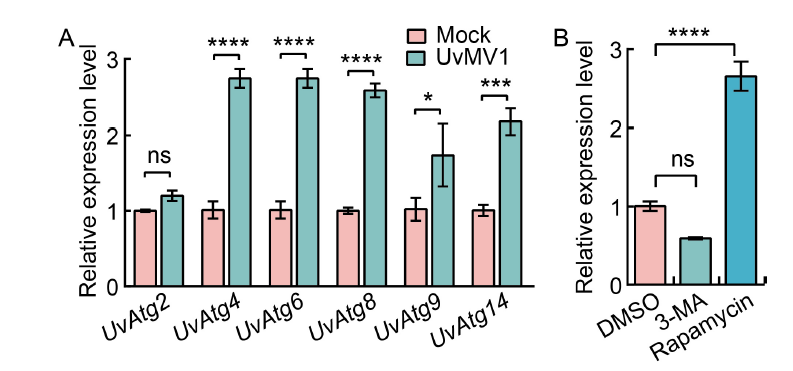
Fig. 3. UvMV1 infection induces autophagy in Ustilaginoide virens to increase viral titers. A, qRT-PCR analysis of the transcript levels of UvAtg2, UvAtg4, UvAtg6, UvAtg8, UvAtg9, and UvAtg14 in mock- and UvMV1-infected U. virens. The mock- and UvMV1-infected strains were cultured in potato sucrose (PS) liquid medium for 5 d with shaking in the dark, after which total RNA was extracted for qRT-PCR analyses. B, qRT-PCR analysis shows the impact of 3-methyladenine (3-MA) and rapamycin on the transcript levels of the UvMV1 RNA-dependent RNA polymerase genes in U. virens. The UvMV1-infected strains were initially cultured in PS medium for 2 d, and then 3-MA or rapamycin was added to PS liquid medium for 24 h, respectively. The UvMV1-infected strains were treated by washing them with ddH2O and then incubated on regular PS medium for 3 d. Dimethylsulfoxide (DMSO)-treated U. virens were used as a control. The levels of UvMV1 RNA transcripts in various samples were individually detected using qRT-PCR. Values represent the mean relative to the mock-treated strains, with the housekeeping gene β-tubulin serving as the internal control. Student’s t-test was used for analyses: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; ns, Not significant.
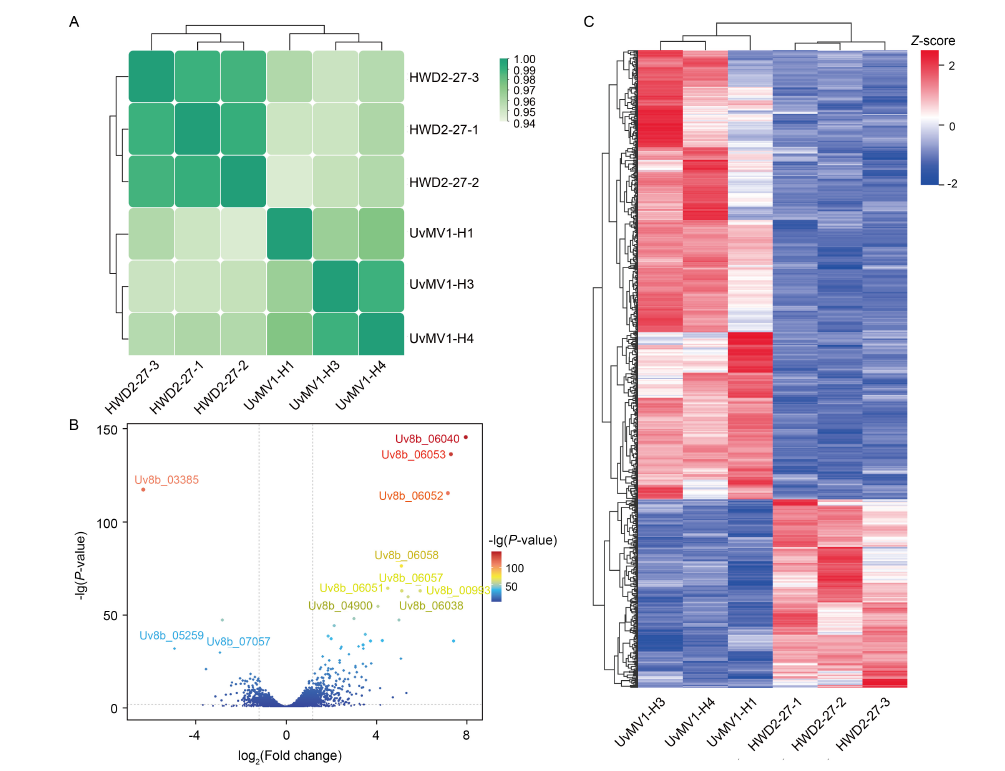
Fig. 4. Overall transcriptomic results and analysis of differentially expressed genes (DEGs) in UvMV1-infected and UvMV1-free strains of Ustilaginoidea virens. A, Sample correlation heatmap. HWD2-27-1, -2, and -3 as well as UvMV1-H1, -3, and -4 are three biological replicates, respectively. B, Volcano plots illustrating differential gene expression levels between UvMV1-infected strains and UvMV1-free strains. FDR, False discovery rate.C, Heatmap illustrating DEGs in UvMV1-infected strains compared with UvMV1-free strains.
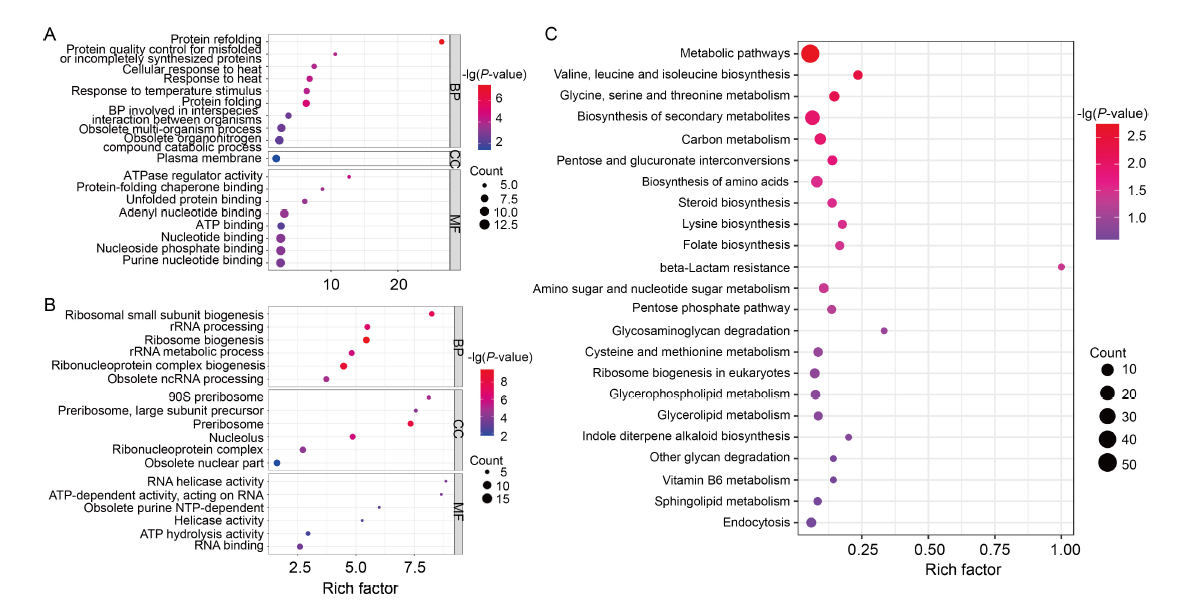
Fig. 5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enhancement analyses of differentially expressed genes (DEGs). A and B, GO terms that are enriched in up-regulated (A) and down-regulated genes (B), as identified through GO enrichment analysis. The bubble size represents the number of members detected in the GO enrichment and the color of the bubble represents the P-value. BP, Biological process; CC, Cellular component; MF, Molecular function.C, KEGG pathway enrichment analysis of DEGs. The rich factor is the ratio of DEG numbers annotated in this pathway term to all gene numbers annotated in this pathway term. The bubble size represents the number of members detected in the KEGG enrichment and the color of the bubble represents the P-value.
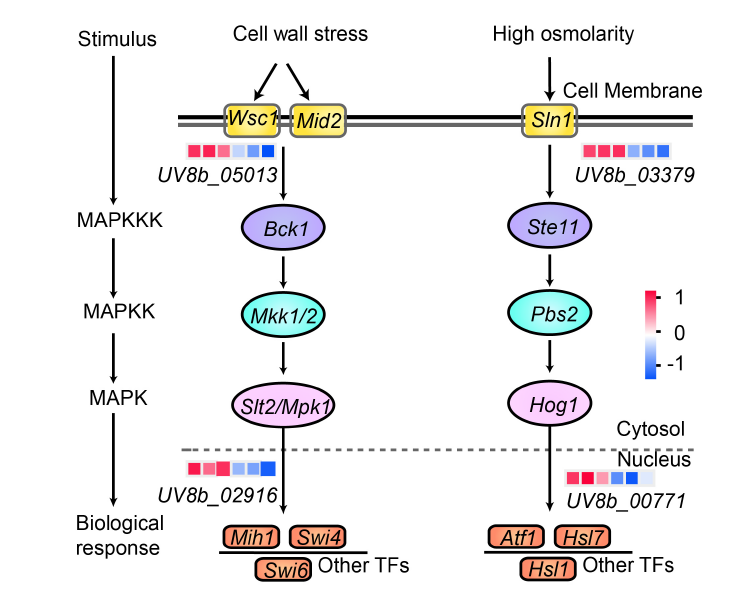
Fig. 6. Differentially expressed genes involved in MAPK signaling pathway. The pathways representing cell wall stress and high osmolarity are illustrated from left to right, respectively. Expression values were presented as log2(Fold change), with red indicating up-regulation and blue indicating down-regulation. In the heatmap, each column represents the expression of HWD2-27-1, HWD2-27-2, HWD2-27-3, UvMV1-H1, UvMV1-H3, and UvMV1-H4 from left to right, and each row represents one gene. MAPK, Mitogen-activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPK kinase kinase; TF, Transcription factor.
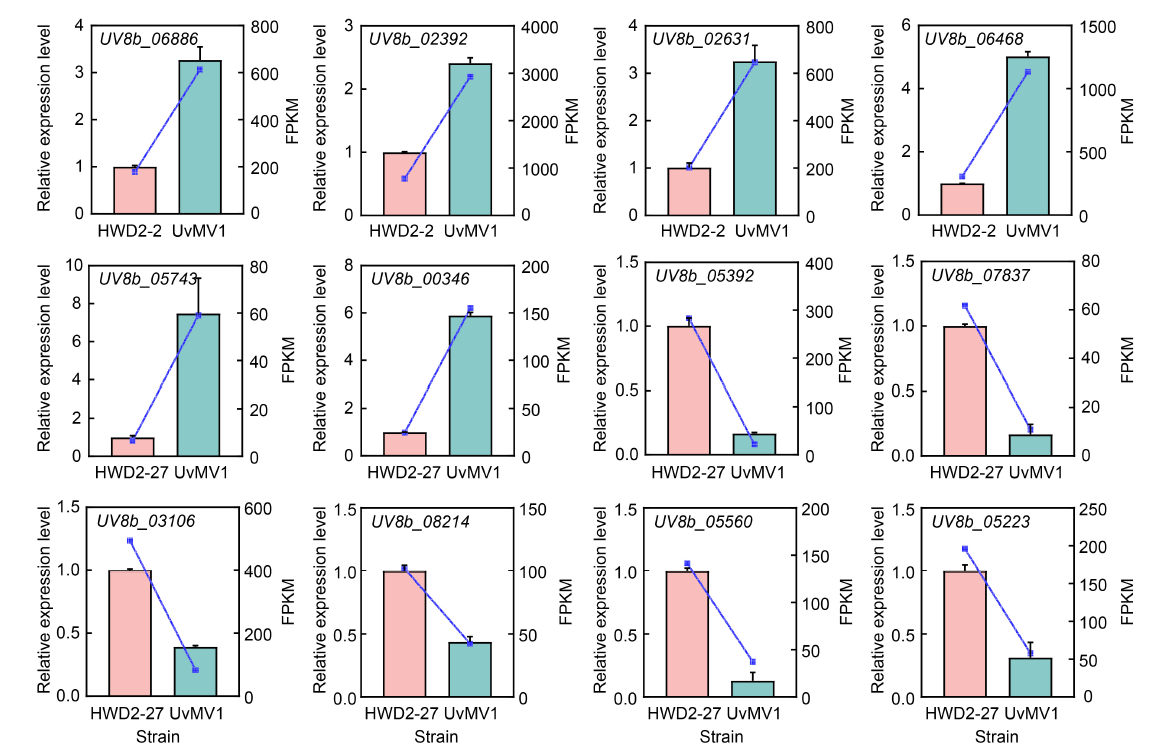
Fig. 7. Accuracy of RNA-seq was validated through qRT-PCR analysis. The expression levels of 12 DEGs identified from the RNA-seq analysis, comprising 6 up-regulated (UV8b_06886, UV8b_02392, UV8b_02631, UV8b_06468, UV8b_05743, and UV8b_00346) and 6 down-regulated genes (UV8b_05392, UV8b_07837, UV8b_03106, UV8b_08214, UV8b_05560, and UV8b_05223), were quantified using qRT-PCR. Histograms were generated from the data obtained via qRT-PCR, while corresponding line charts were constructed from the fragments per kilobase of transcript per million mapped reads (FPKM) values derived from the RNA-seq analysis, with distinct colors representing different samples. Each bar in the histograms denotes the mean and standard error of three independent experiments.
| [1] | Buck K W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res, 47: 159-251. |
| [2] | Byun J Y, Yoon C H, An S, et al. 2009. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis, 30(11): 1880-1888. |
| [3] | Chen X Y, Pei Z X, Liu H, et al. 2022. Host-induced gene silencing of fungal-specific genes of Ustilaginoidea virens confers effective resistance to rice false smut. Plant Biotechnol J, 20(2): 253-255. |
| [4] | Chen Y H, Du W L, Hagemeijer M C, et al. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell, 160(4): 619-630. |
| [5] | Chun S J, Lee Y H. 1997. Inheritance of dsRNAs in the rice blast fungus, Magnaporthe grisea. FEMS Microbiol Lett, 148(2): 159-162. |
| [6] | Cooke M C. 1878. Some extra-European fungi. Grevillea, 7: 13-15. |
| [7] | Ding S W. 2010. RNA-based antiviral immunity. Nat Rev Immunol, 10: 632-644. |
| [8] | Donaire L, Ayllón M A. 2017. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol Plant Pathol, 18(8): 1127-1137. |
| [9] | Fan J, Guo X Y, Li L, et al. 2015. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes. J Integr Plant Biol, 57(6): 577-590. |
| [10] | Fan J, Du N, Li L, et al. 2019. A core effector UV_1261 promotes Ustilaginoidea virens infection via spatiotemporally suppressing plant defense. Phytopathol Res, 1(1): 11. |
| [11] | Fan J, Liu J, Gong Z Y, et al. 2020. The false smut pathogen Ustilaginoidea virens requires rice stamens for false smut ball formation. Environ Microbiol, 22(2): 646-659. |
| [12] | Fan Y, Zhao W H, Tang X L, et al. 2024. Co-infection of four novel mycoviruses from three lineages confers hypovirulence on phytopathogenic fungus Ustilaginoidea virens. Rice, 17(1): 44. |
| [13] | Ghabrial S A, Castón J R, Jiang D H, et al. 2015. 50-plus years of fungal viruses. Virology, 479/480: 356-368. |
| [14] | Grente J, Berthelay-Sauret S. 1978. Biological control of chestnut blight in France. In: Proceedings of the American Chestnut Symposium. January 4-5, 1978. Morgantown, West Virginia, USA: West Virginia University Press. |
| [15] | Hai D, Li J C, Jiang D H, et al. 2024. Plants interfere with non-self recognition of a phytopathogenic fungus via proline accumulation to facilitate mycovirus transmission. Nat Commun, 15(1): 4748. |
| [16] | He Z R, Huang X T, Fan Y, et al. 2022. Metatranscriptomic analysis reveals rich mycoviral diversity in three major fungal pathogens of rice. Int J Mol Sci, 23(16): 9192. |
| [17] | Hickey L T, Hafeez A N, Robinson H, et al. 2019. Breeding crops to feed 10 billion. Nat Biotechnol, 37(7): 744-754. |
| [18] | Hollings M. 1962. Viruses associated with a die-back disease of cultivated mushroom. Nature, 196: 962-965. |
| [19] | Huang X Q, Chen S P, Yang X R, et al. 2020. Friend or enemy: A dual role of autophagy in plant virus infection. Front Microbiol, 11: 736. |
| [20] | Huang X Q, Wang J K, Chen S P, et al. 2024. Rhabdovirus encoded glycoprotein induces and harnesses host antiviral autophagy for maintaining its compatible infection. Autophagy, 20(2): 275-294. |
| [21] | Khalifa M E, Pearson M N. 2013. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology, 441(1): 22-30. |
| [22] | Kondo H, Botella L, Suzuki N. 2022. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu Rev Phytopathol, 60: 307-336. |
| [23] | Lakshman D K. 1994. Spontaneous appearance of genetically distinct double-stranded RNA elements in Rhizoctonia solani. Phytopathology, 84: 633-639. |
| [24] | Li F F, Zhang C W, Li Y Z, et al. 2018. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat Commun, 9(1): 1268. |
| [25] | Li P F, Wang S C, Zhang L H, et al. 2020. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci Adv, 6(14): eaay9634. |
| [26] | Liu S, Xie J T, Cheng J S, et al. 2016. Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc Natl Acad Sci USA, 113(45): 12803-12808. |
| [27] | Lu M H, Liu W C, Zhu F. 2018. Discussion on epidemic rule of rice false smut in recent years and its controlling strategy. China Plant Prot, 38(5): 44-47. (in Chinese with English abstract) |
| [28] | Morris T J, Dodds J A. 1979. Isolation and analysis of double- stranded RNA from virus-infected plant and fungal tissue. Phytopathology, 69(8): 854-858. |
| [29] | Muñoz-Adalia E J, Diez J J, Fernández M M, et al. 2018. Characterization of small RNAs originating from mitoviruses infecting the conifer pathogen Fusarium circinatum. Arch Virol, 163(4): 1009-1018. |
| [30] | Nibert M L. 2017. Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology, 507: 96-100. |
| [31] | Nibert M L, Vong M, Fugate K K, et al. 2018. Evidence for contemporary plant mitoviruses. Virology, 518: 14-24. |
| [32] | Polashock J J, Hillman B I. 1994. A small mitochondrial double- stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc Natl Acad Sci USA, 91(18): 8680-8684. |
| [33] | Ren J Y, Zhang Y H, Wang Y H, et al. 2022. Deletion of all three MAP kinase genes results in severe defects in stress responses and pathogenesis in Fusarium graminearum. Stress Biol, 2(1): 6. |
| [34] | Rigling D, Prospero S. 2018. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol Plant Pathol, 19(1): 7-20. |
| [35] | Romeralo C, Bezos D, Martínez-Álvarez P, et al. 2018. Vertical transmission of Fusarium circinatum mitoviruses FcMV1 and FcMV2-2 via microconidia. Forests, 9(6): 356. |
| [36] | Saccardo F, Cettul E, Palmano S, et al. 2011. On the alleged origin of geminiviruses from extrachromosomal DNAs of phytoplasmas. BMC Evol Biol, 11: 185. |
| [37] | Samsa M M, Mondotte J A, Iglesias N G, et al. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog, 5(10): e1000632. |
| [38] | Shafik K, Umer M, You H F, et al. 2021. Characterization of a novel mitovirus infecting Melanconiella theae isolated from tea plants. Front Microbiol, 12: 757556. |
| [39] | Shahi S, Eusebio-Cope A, Kondo H, et al. 2019. Investigation of host range of and host defense against a mitochondrially replicating mitovirus. J Virol, 93(6): e01503-18. |
| [40] | Sir D, Ou J H J. 2010. Autophagy in viral replication and pathogenesis. Mol Cells, 29(1): 1-8. |
| [41] | Song J H, Wei W, Lv B, et al. 2016. Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ Microbiol, 18(11): 3840-3849. |
| [42] | Sun W X, Fan J, Fang A F, et al. 2020. Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu Rev Phytopathol, 58: 363-385. |
| [43] | Turina M, Ghignone S, Astolfi N, et al. 2018. The virome of the arbuscular mycorrhizal fungus Gigaspora margarita reveals the first report of DNA fragments corresponding to replicating non- retroviral RNA viruses in fungi. Environ Microbiol, 20(6): 2012-2025. |
| [44] | Vainio E J, Jurvansuu J, Streng J, et al. 2015. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J Gen Virol, 96(Pt 3): 714-725. |
| [45] | Vainio E J, Jurvansuu J, Hyder R, et al. 2018. Heterobasidion partitivirus 13 mediates severe growth debilitation and major alterations in the gene expression of a fungal forest pathogen. J Virol, 92(5): e01744-17. |
| [46] | Walker P J, Siddell S G, Lefkowitz E J, et al. 2022. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch Virol, 167(11): 2429-2440. |
| [47] | Wang J, Xu C Y, Sun Q M, et al. 2021. Post-translational regulation of autophagy is involved in intra-microbiome suppression of fungal pathogens. Microbiome, 9(1): 131. |
| [48] | Wang Q, Kawano Y. 2022. Improving disease resistance to rice false smut without yield penalty by manipulating the expression of effector target. Mol Plant, 15(12): 1834-1837. |
| [49] | Wang Q, Lu L N, Zeng M, et al. 2022. Rice black-streaked dwarf virus P 10 promotes phosphorylation of GAPDH (glyceraldehyde- 3-phosphate dehydrogenase) to induce autophagy in Laodelphax striatellus. Autophagy, 18(4): 745-764. |
| [50] | Wu M D, Zhang L, Li G Q, et al. 2007. Hypovirulence and double- stranded RNA in Botrytis cinerea. Phytopathology, 97(12): 1590-1599. |
| [51] | Xie J T, Ghabrial S A. 2012. Molecular characterizations of two mitoviruses co-infecting a hyovirulent isolate of the plant pathogenic fungus Sclerotinia sclerotiorum. Virology, 428(2): 77-85. |
| [52] | Xie J T, Jiang D H. 2014. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol, 52: 45-68. |
| [53] | Xie S W, Shi H B, Wen H, et al. 2024. Carbon catabolite repressor UvCreA is required for development and pathogenicity in Ustilaginoidea virens. Rice Sci, 31(2): 203-214. |
| [54] | Xu Z Y, Wu S S, Liu L J, et al. 2015. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res, 197: 127-136. |
| [55] | Yang M, Zhang Y L, Xie X L, et al. 2018. Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell, 30(7): 1582-1595. |
| [56] | Yang M, Ismayil A, Liu Y L. 2020. Autophagy in plant-virus interactions. Annu Rev Virol, 7(1): 403-419. |
| [57] | Yu X, Li B, Fu Y P, et al. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA, 107(18): 8387-8392. |
| [58] | Yu X, Li B, Fu Y P, et al. 2013. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci USA, 110(4): 1452-1457. |
| [59] | Zhang H X, Xie J T, Fu Y P, et al. 2020. A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for Brassica protection and yield enhancement. Mol Plant, 13(10): 1420-1433. |
| [60] | Zhang L, Liu W W, Wu N, et al. 2023. Southern rice black-streaked dwarf virus induces incomplete autophagy for persistence in gut epithelial cells of its vector insect. PLoS Pathog, 19(1): e1011134. |
| [61] | Zheng L, Liu C, Zhang M L, et al. 2018. Diversity of dsRNA viruses infecting rice sheath blight fungus Rhizoctonia solani AG-1 IA. Rice Sci, 25(1): 57-60. |
| [62] | Zhu J Z, Li P, Zhang Z, et al. 2024. The CfKOB1 gene related to cell apoptosis is required for pathogenicity and involved in mycovirus-induced hypovirulence in Colletotrichum fructicola. Int J Biol Macromol, 271(Pt 1): 132437. |
| [1] | Zhang Fengmin, Cao Zhenzhen, Zheng Xin, He Yuntao, Chen Mingxue, Lin Xiaoyan. Interaction Between Ustilaginoidea virens and Rice and Its Sustainable Control [J]. Rice Science, 2024, 31(3): 269-284. |
| [2] | Xie Shuwei, Shi Huanbin, Wen Hui, Liu Zhiquan, Qiu Jiehua, Jiang Nan, Kou Yanjun. Carbon Catabolite Repressor UvCreA is Required for Development and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2024, 31(2): 203-214. |
| [3] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| [4] | Wei Qinghui, Cui Daizong, Zheng Baojiang, Zhao Min. In Vitro Antifungal Activity of Dihydrochelerythrine and Proteomic Analysis in Ustilaginoidea virens [J]. Rice Science, 2023, 30(3): 257-266. |
| [5] | Liu Yueran, Qu Jinsong, Wang Yufu, Yin Weixiao, Luo Chaoxi. bZIP Transcription Factor UvATF21 Mediates Vegetative Growth, Conidiation, Stress Tolerance and Is Required for Full Virulence of Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2023, 30(1): 50-57. |
| [6] | Meng Shuai, Qiu Jiehua, Xiong Meng, Liu Zhiquan, Jane Sadhna Jagernath, Lin Fucheng, Shi Huanbin, Kou Yanjun. UvWhi2 Is Required for Stress Response and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2022, 29(1): 47-54. |
| [7] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [8] | Junjie Yu, Mina Yu, Tianqiao Song, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Xiaole Yin, Dong Liang, Yongfeng Liu. UvSMEK1, a Suppressor of MEK Null, Regulates Pathogenicity, Conidiation and Conidial Germination in Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2021, 28(5): 457-465. |
| [9] | Meng Xiong, Shuai Meng, Jiehua Qiu, Huanbin Shi, Xiangling Shen, Yanjun Kou. Putative Phosphatase UvPsr1 Is Required for Mycelial Growth, Conidiation, Stress Response and Pathogenicity in Ustilaginonidea virens [J]. Rice Science, 2020, 27(6): 529-536. |
| [10] | Jiehua Qiu, Shuai Meng, Yizhen Deng, Shiwen Huang, Yanjun Kou. Ustilaginoidea virens: A Fungus Infects Rice Flower and Threats World Rice Production [J]. Rice Science, 2019, 26(4): 199-206. |
| [11] | Palaniyandi VELUSAMY, J. EBENEZAR IMMANUEL, Samuel,S. GNANAMANICKAM. Rhizosphere Bacteria for Biocontrol of Bacterial Blight and Growth Promotion of Rice [J]. RICE SCIENCE, 2013, 20(5): 356-362. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||