
Rice Science ›› 2020, Vol. 27 ›› Issue (6): 529-536.DOI: 10.1016/j.rsci.2020.09.009
• Research Paper • Previous Articles
Meng Xiong1,2, Shuai Meng2, Jiehua Qiu2, Huanbin Shi2, Xiangling Shen1( ), Yanjun Kou2(
), Yanjun Kou2( )
)
Received:2019-12-03
Accepted:2020-05-09
Online:2020-11-28
Published:2020-11-28
About author:#These authors contributed equally to this work
Meng Xiong, Shuai Meng, Jiehua Qiu, Huanbin Shi, Xiangling Shen, Yanjun Kou. Putative Phosphatase UvPsr1 Is Required for Mycelial Growth, Conidiation, Stress Response and Pathogenicity in Ustilaginonidea virens[J]. Rice Science, 2020, 27(6): 529-536.
Add to citation manager EndNote|Ris|BibTeX
| Primer | Sequence (5′-3′) | Application |
|---|---|---|
| UvPSR1-3F | AAAACTGCAGTGCCAAAATTACCCGACGGT | Amplifying UvPSR1 3' flank sequence for gene deletion |
| UvPSR1-3R | AAAACTGCAGGGTACGGTGCACACGAAGTA | |
| UvPSR1-5F | CCGCTCGAGCCCAGAGTTGTAGTCGGCTG | Amplifying UvPSR1 5' flank sequence for gene deletion |
| UvPSR1-5R | CGGGATCCCCTTTGGAAGTCCCCACCTC | |
| UvPSR1-TR | CGGGCTCACTCCTGATTCTT | Transformants screening |
| p821-3F | CAGCACTCGTCCGAGGGCA | |
| UvPSR1-CF UvPSR1-CR UvPSR1RT-F UvPSR1RT-R | CGGAATTCGCATCGTCCACGAATCTAGCTGTAG GCTCTAGAGCAACAACCTTCGTAATTCCTCGC CGTTGAGATCGAGGGCCATT CCGGAAGAGACGGTGATGTA | Uvpsr1Δ complementation assay Analysis the expression level of UvPSR1 gene of U. virens |
| β-tubulin F | GGCGTTTACAATGGCACTTC | Analysis the expression level of β-tubulin gene of U. virens |
| β-tubulin R | CGGAACAGTTGACCAAAAGG | |
| UvPSR1-probeF | CCGCTCGAGCCCAGAGTTGTAGTCGGCTG | Amplification of probe for Southern Blot assay Amplification of neomycin resistance gene to construct the vector pFGL823 |
| UvPSR1-probeR Neo-p823-F Neo-p823-R | CGGGATCCCCTTTGGAAGTCCCCACCTC CTAGTCTAGAGATTAACGCTTACAATTTCCATTCG AAAACTGCAGAGAATAGGAACTTCGGAATAGG |
Supplemental Table 1. Primers used in this study.
| Primer | Sequence (5′-3′) | Application |
|---|---|---|
| UvPSR1-3F | AAAACTGCAGTGCCAAAATTACCCGACGGT | Amplifying UvPSR1 3' flank sequence for gene deletion |
| UvPSR1-3R | AAAACTGCAGGGTACGGTGCACACGAAGTA | |
| UvPSR1-5F | CCGCTCGAGCCCAGAGTTGTAGTCGGCTG | Amplifying UvPSR1 5' flank sequence for gene deletion |
| UvPSR1-5R | CGGGATCCCCTTTGGAAGTCCCCACCTC | |
| UvPSR1-TR | CGGGCTCACTCCTGATTCTT | Transformants screening |
| p821-3F | CAGCACTCGTCCGAGGGCA | |
| UvPSR1-CF UvPSR1-CR UvPSR1RT-F UvPSR1RT-R | CGGAATTCGCATCGTCCACGAATCTAGCTGTAG GCTCTAGAGCAACAACCTTCGTAATTCCTCGC CGTTGAGATCGAGGGCCATT CCGGAAGAGACGGTGATGTA | Uvpsr1Δ complementation assay Analysis the expression level of UvPSR1 gene of U. virens |
| β-tubulin F | GGCGTTTACAATGGCACTTC | Analysis the expression level of β-tubulin gene of U. virens |
| β-tubulin R | CGGAACAGTTGACCAAAAGG | |
| UvPSR1-probeF | CCGCTCGAGCCCAGAGTTGTAGTCGGCTG | Amplification of probe for Southern Blot assay Amplification of neomycin resistance gene to construct the vector pFGL823 |
| UvPSR1-probeR Neo-p823-F Neo-p823-R | CGGGATCCCCTTTGGAAGTCCCCACCTC CTAGTCTAGAGATTAACGCTTACAATTTCCATTCG AAAACTGCAGAGAATAGGAACTTCGGAATAGG |
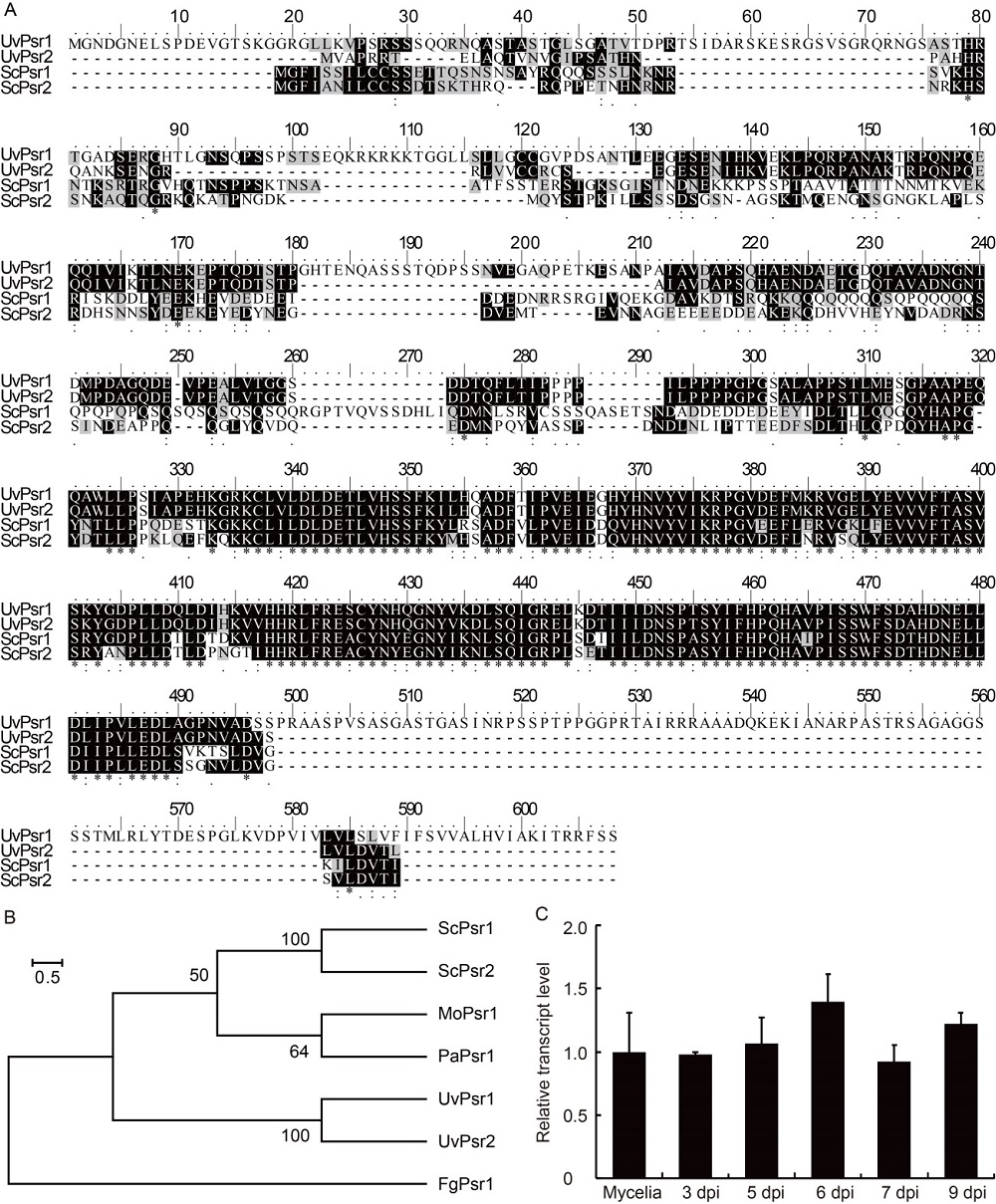
Fig. 1. Identification of UvPsr1 in Ustilaginonidea virens. A, Alignment of Psr1 proteins produced by U. virens (UvPsr1 and UvPsr2) and Saccharomyces cerevisiae (ScPsr1 and ScPsr2). Amino acids in black and gray represent amino acids identity and 50% similairty respectively. B, Phylogeny of the Psr proteins, derived from an alignment of homologous proteins encoded by U. virens (UvPsr1), S. cerevisiae (ScPsr1), Magnaporthe oryzae (MoPsr1), Podospora anserina (PaPsr1) and Fusarium graminearum (FgPsr1). C, Abundance of UvPSR1 transcript following the inoculation of rice panicles with U. virens. Samples were taken at 3, 5, 6, 7 and 9 dpi (days post inoculation). β-tubulin gene served as the reference. Data are shown as Mean ± SD (n = 3).
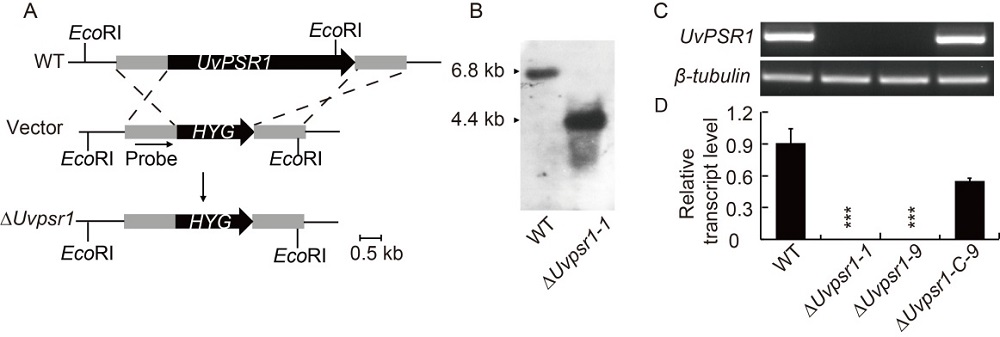
Fig. 2. Targeted gene deletion and complementation of UvPSR1 in U. virens. A, Strategy for constructing a ∆Uvpsr1 strain (WT) involved replacing UvPSR1 with hygromycin phosphotransferase gene cassette (HYG). B, Southern blot assay used to validate the loss of UvPSR1 in the ∆Uvpsr1 deletion mutant. C and D, Abundance of UvPSR1 transcript in mycelia of the WT, ∆Uvpsr1 and ∆Uvpsr1-C strains. No transcript was detected in either the ∆Uvpsr1-1 or ∆Uvpsr1-9 strains. Data are shown as Mean ± SD (n = 3). *** indicates the significant difference at the 0.001 level.
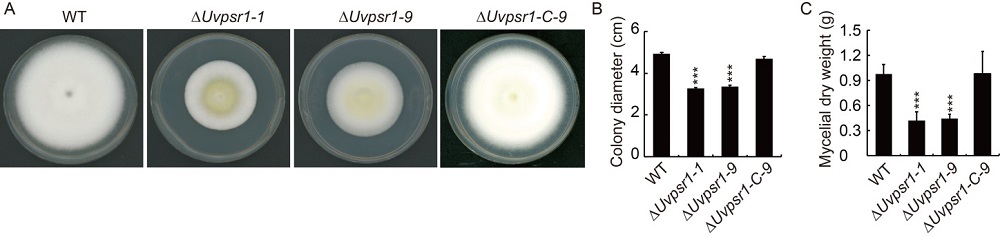
Fig. 3. UvPsr1 is needed for mycelial growth of U. virens. A, Colonies of the WT, ∆Uvpsr1 and ∆Uvpsr1-C strains grown on potato sucrose agar (PSA) in the dark for 15 d. B, Quantification of fungal growth as measured by colony diameter. C, Dry weight of mycelia developed in potato sucrose medium for 7 d. Data are shown as Mean ± SD (n = 3). *** indicates significant difference at the 0.001 level.
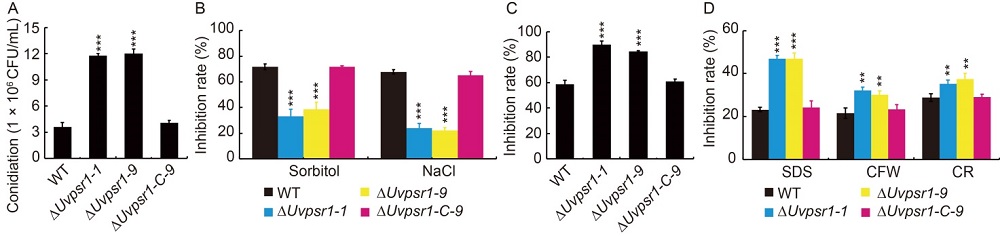
Fig. 4. Effects of UvPSR1 in production of conidia (A), sensitivity to osmosis and salinity stresses (B), tolerance to oxidative stress (C), and tolerance to cell wall disruption induced by SDS (sodium dodecyl sulfate), CFW (calcofluor white stain) and CR (congo red) (D).Data are shown as Mean ± SD (n = 3). ** and *** indicate significant differences at the 0.01 and 0.001 levels, respectively.
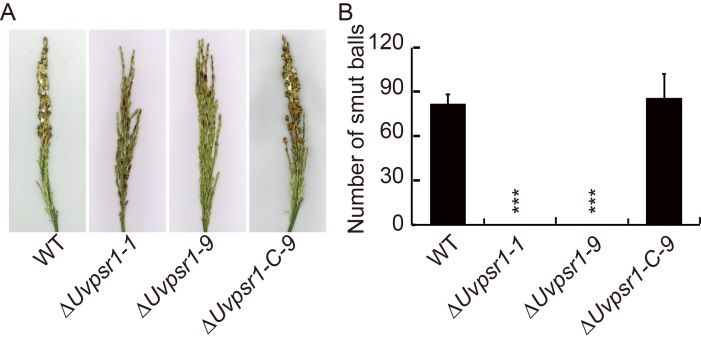
Fig. 5. Pathogenicity of U. virens requires the presence of UvPsr1. A, Disease symptoms on the rice panicle following inoculation at the booting stage with a mixture of mycelia and conidia of either the WT, ∆Uvpsr1 or ∆Uvpsr1-C-9 strains. B, Quantification of pathogenicity as measured by the number of smut balls formed within the panicle. Data are shown as Mean ± SD (n = 3). *** indicates significant difference at the 0.001 level.
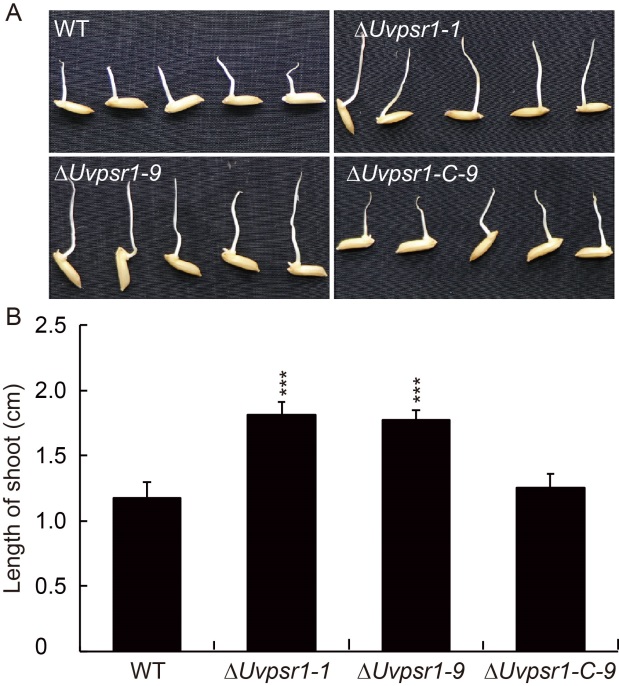
Fig. 6. Culture filtrates of ∆Uvpsr1 mutants showed reduced inhibition of rice seed elongation. A, Grains were germinated in the presence of the filtrate of WT strain, UvPSR1 mutants (∆Uvpsr1-1 and ∆Uvpsr1-9) and complementary (∆Uvpsr1-C-9) strain. B, Quantification of shoot lengths. Data are shown as Mean ± SD (n = 3). *** indicates the significant difference at the 0.001 level.
| [1] | Ashizawa T, Takahashi M, Moriwaki J, Hirayae K.2010. A refined inoculation method to evaluate false smut resistance in rice. J Gen Plant Pathol, 77(1): 10-16. |
| [2] | Boeckstaens M, Llinares E, van Vooren P, Marini A M.2014. The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nat Commun, 5: 3101. |
| [3] | Chen X Y, Hai D, Tang J T, Liu H, Huang J B, Luo C X, Hsiang T, Zheng L.2020. UvCom1 is an important regulator required for development and infection in the rice false smut fungus Ustilaginoidea virens. Phytopathology, 110(2): 483-493. |
| [4] | Chen Y Y, Stabryla L, Wei N.2016. Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Appl Environ Microbiol, 82(7): 2156-2166. |
| [5] | Fan J, Yang J, Wang Y Q, Li G B, Li Y, Huang F, Wang W M.2016. Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease. Mol Plant Pathol, 17(9): 1321-1330. |
| [6] | Fan J, Du N, Li L, Li G B, Wang Y Q, Zhou Y F, Hu X H, Liu J, Zhao J Q, Li Y, Huang F, Wang W M.2019. A core effector UV_1261 promotes Ustilaginoidea virens infection via spatiotemporally suppressing plant defense. Phytopathol Res, 1(1): 11. |
| [7] | Fang A F, Gao H, Zhang N, Zheng X H, Qiu S S, Li Y J, Zhou S, Cui F H, Sun W X.2019. A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Front Microbiol, 10: 845. |
| [8] | Guo W W, Gao Y X, Yu Z M, Xiao Y H, Zhang Z G, Zhang H F.2019. The adenylate cyclase UvAc1 and phosphodiesterase UvPdeH control the intracellular cAMP level, development, and pathogenicity of the rice false smut fungus Ustilaginoidea virens. Fungal Genet Biol, 129: 65-73. |
| [9] | Kaida D, Yashiroda H, Toh-e A, Kikuchi Y.2002. Yeast Whi2 and Psr1-phosphatase form a complex and regulate STRE-mediated gene expression. Genes Cells, 7(6): 543-552. |
| [10] | Ladhalakshmi D, Laha G S, Singh R, Karthikeyan A, Mangrauthia S K, Sundaram R M, Thukkaiyannan P, Viraktamath B C.2012. Isolation and characterization of Ustilaginoidea virens and survey of false smut disease of rice in India. Phytoparasitica, 40(2): 171-176. |
| [11] | Liang Y F, Han Y, Wang C F, Jiang C, Xu J R.2018. Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front Plant Sci, 9: 699. |
| [12] | Lin X Y, Bian Y F, Mou R X, Cao Z Y, Cao Z Z, Zhu Z W, Chen M X.2018. Isolation, identification, and characterization of Ustilaginoidea virens from rice false smut balls with high ustilotoxin production potential. J Basic Microbiol, 58(8): 670-678. |
| [13] | Lv B, Zheng L, Liu H, Tang J T, Hsiang T, Huang J B.2016. Use of random T-DNA mutagenesis in identification of gene UvPRO1, a regulator of conidiation, stress response, and virulence in Ustilaginoidea virens. Front Microbiol, 7: 2086. |
| [14] | Nessa B, Salam M U, Haque A H M M, Biswas J K, Kabir M S, MacLeod W J, D’Antuono M, Barman H N, Latif M A, Galloway J.2015. Spatial pattern of natural spread of rice false smut Ustilaginoidea virens disease in fields. Am J Agric Biol Sci, 10(2): 63-73. |
| [15] | Qiu J H, Meng S, Deng Y Z, Huang S W, Kou Y J.2019. Ustilaginoidea virens: A fungus infects rice flower and threats world rice production. Rice Sci, 26(4): 199-206. |
| [16] | Tang J T, Bai J, Chen X Y, Zheng L, Liu H, Huang J B.2019. Two protein kinases UvPmk1 and UvCDC2 with significant functions in conidiation, stress response and pathogenicity of rice false smut fungus Ustilaginoidea virens. Curr Genet, 66: 409-420. |
| [17] | Teng X C, Hardwick J M.2019. Whi2: A new player in amino acid sensing. Curr Genet, 65(3): 701-709. |
| [18] | Timpano H, Tong L C H, Gautier V, Lalucque H, Silar P.2016. The PaPsr1 and PaWhi2 genes are members of the regulatory network that connect stationary phase to mycelium differentiation and reproduction in Podospora anserina. Fungal Genet Biol, 94: 1-10. |
| [19] | Wang X H, Wang J, Lai D W, Wang W X, Dai J G, Zhou L G, Liu Y.2017. Ustiloxin G, a new cyclopeptide mycotoxin from rice false smut balls. Toxins, 9(2): E54. |
| [20] | Xie S L, Wang Y F, Wei W, Li C Y, Liu Y, Qu J S, Meng Q H, Lin Y, Yin W X, Yang Y N, Luo C X.2019. The Bax inhibitor UvBI-1, a negative regulator of mycelial growth and conidiation, mediates stress response and is critical for pathogenicity of the rice false smut fungus Ustilaginoidea virens. Curr Genet, 65(5): 1185-1197. |
| [21] | Yu M N, Yu J J, Hu J K, Huang L, Wang Y H, Yin X L, Nie Y F, Meng X K, Wang W D, Liu Y F.2015. Identification of pathogenicity-related genes in the rice pathogen Ustilaginoidea virens through random insertional mutagenesis. Fungal Genet Biol, 76: 10-19. |
| [22] | Yun Y Z, Liu Z Y, Yin Y N, Jiang J H, Chen Y, Xu J R, Ma Z H.2015. Functional analysis of the Fusarium graminearum phosphatome. New Phytol, 207(1): 119-134. |
| [23] | Zhang Y, Zhang K, Fang A F, Han Y Q, Yang J, Xue M F, Bao J D, Hu D W, Zhou B, Sun X Y, Li S J, Wen M, Yao N, Ma L J, Liu Y F, Zhang M, Huang F, Luo C X, Zhou L G, Li J Q, Chen Z Y, Miao J K, Wang S, Lai J S, Xu J R, Hsiang T, Peng Y L, Sun W X.2014. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun, 5: 3849. |
| [24] | Zheng D W, Wang Y, Han Y, Xu J R, Wang C F.2016. UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci Rep, 6: 24824. |
| [25] | Zheng M T, Ding H, Huang L, Wang Y H, Yu M N, Zheng R, Yu J J, Liu Y F.2017. Low-affinity iron transport protein Uvt3277 is important for pathogenesis in the rice false smut fungus Ustilaginoidea virens. Curr Genet, 63(1): 131-144. |
| [26] | Zhou Y X, Yu J J, Pan X Y, Yu M N, Du Y, Qi Z Q, Zhang R S, Song T Q, Yin X L, Liu Y F.2019. Characterization of propiconazole field-resistant isolates of Ustilaginoidea virens. Pestic Biochem Physiol, 153: 144-151. |
| [1] | Wei Qinghui, Cui Daizong, Zheng Baojiang, Zhao Min. In Vitro Antifungal Activity of Dihydrochelerythrine and Proteomic Analysis in Ustilaginoidea virens [J]. Rice Science, 2023, 30(3): 257-266. |
| [2] | Liu Yueran, Qu Jinsong, Wang Yufu, Yin Weixiao, Luo Chaoxi. bZIP Transcription Factor UvATF21 Mediates Vegetative Growth, Conidiation, Stress Tolerance and Is Required for Full Virulence of Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2023, 30(1): 50-57. |
| [3] | Meng Shuai, Qiu Jiehua, Xiong Meng, Liu Zhiquan, Jane Sadhna Jagernath, Lin Fucheng, Shi Huanbin, Kou Yanjun. UvWhi2 Is Required for Stress Response and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2022, 29(1): 47-54. |
| [4] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [5] | Junjie Yu, Mina Yu, Tianqiao Song, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Xiaole Yin, Dong Liang, Yongfeng Liu. UvSMEK1, a Suppressor of MEK Null, Regulates Pathogenicity, Conidiation and Conidial Germination in Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2021, 28(5): 457-465. |
| [6] | Jiehua Qiu, Shuai Meng, Yizhen Deng, Shiwen Huang, Yanjun Kou. Ustilaginoidea virens: A Fungus Infects Rice Flower and Threats World Rice Production [J]. Rice Science, 2019, 26(4): 199-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||