
Rice Science ›› 2022, Vol. 29 ›› Issue (4): 340-352.DOI: 10.1016/j.rsci.2021.11.005
• Research Paper • Previous Articles Next Articles
Kittisak Buddhachat1( ), Nattaporn Sripairoj1, Onchira Ritbamrung1, Phithak Inthima1, Kumrop Ratanasut2,3, Thanita Boonsrangsom2,3, Tepsuda Rungrat2,3, Pongsanat Pongcharoen2,3, Kawee Sujipuli2,3(
), Nattaporn Sripairoj1, Onchira Ritbamrung1, Phithak Inthima1, Kumrop Ratanasut2,3, Thanita Boonsrangsom2,3, Tepsuda Rungrat2,3, Pongsanat Pongcharoen2,3, Kawee Sujipuli2,3( )
)
Received:2021-09-06
Accepted:2021-11-08
Online:2022-07-28
Published:2022-05-12
Contact:
Kittisak Buddhachat, Kawee Sujipuli
Kittisak Buddhachat, Nattaporn Sripairoj, Onchira Ritbamrung, Phithak Inthima, Kumrop Ratanasut, Thanita Boonsrangsom, Tepsuda Rungrat, Pongsanat Pongcharoen, Kawee Sujipuli. RPA-Assisted Cas12a System for Detecting Pathogenic Xanthomonas oryzae, a Causative Agent for Bacterial Leaf Blight Disease in Rice[J]. Rice Science, 2022, 29(4): 340-352.
Add to citation manager EndNote|Ris|BibTeX
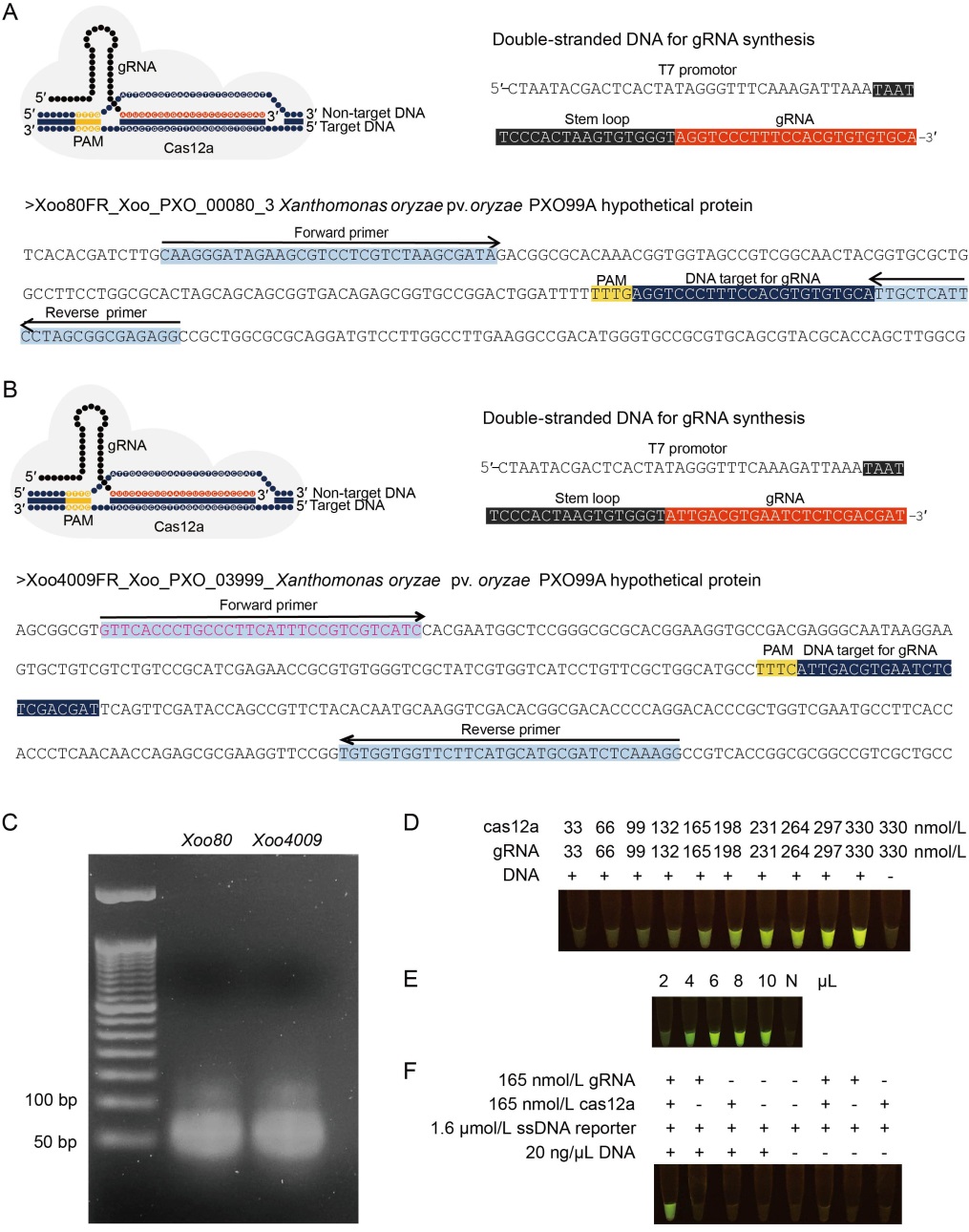
Fig. 1. Guide RNA (gRNA) synthesis and in vitro cas12a digestion. A and B, Tertiary complexes for cas12a: gRNA:DNA targets, DNA templates for gRNA synthesis using in vitro transcription with T7 RNA polymerase and DNA targets containing gRNA binding sites for Xoo80 (A) and Xoo4009 (B) were amplified by recombinase polymerase amplification (RPA). Protospacer adjacent motif (PAM) was used as a proto-adjacent motif. C, Synthesized gRNA-Xoo80 and gRNA- Xoo4009 were visualized under 3% agarose gel electrophoresis. D?F, Experimental procedures were conducted under in vitro digestion to assess the efficiency of concentration ratios of cas12a and gRNA varied from 33:33 to 330:330 nmol/L. E, Sensitivity of various amounts of RPA-Xoo4009 product ranging from 2 to 10 µL. N represents negative control (without RPA product). F, dsDNA digestibility with different chemical modifications (gRNA-Xoo4009, cas12a and RPA-Xoo4009 product).
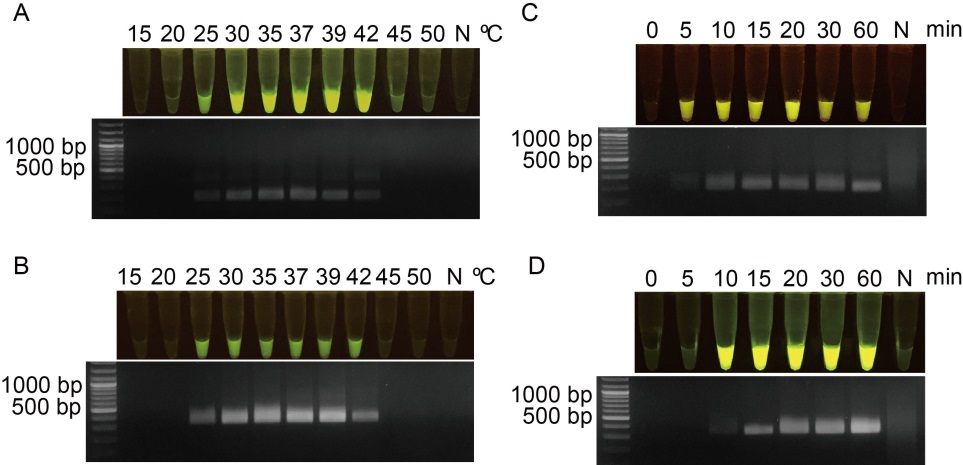
Fig. 2. Optimization of recombinase polymerase amplification (RPA) reaction for RPA-DNA amplification based on Xoo80 or Xoo4009 primer pairs. A and B, RPA was performed under various temperatures ranging from 15 ºC to 50 ºC for RPA-Xoo80 (A) and RPA-Xoo4009 (B) primer pairs. C and D, Time duration for DNA amplification was conducted at different periods of 0?60 min for RPA-Xoo80 (C) and RPA-Xoo4009 (D) primer pairs after obtaining the optimal temperature of 39 ºC. N represents negative control which was the reaction without DNA templates.
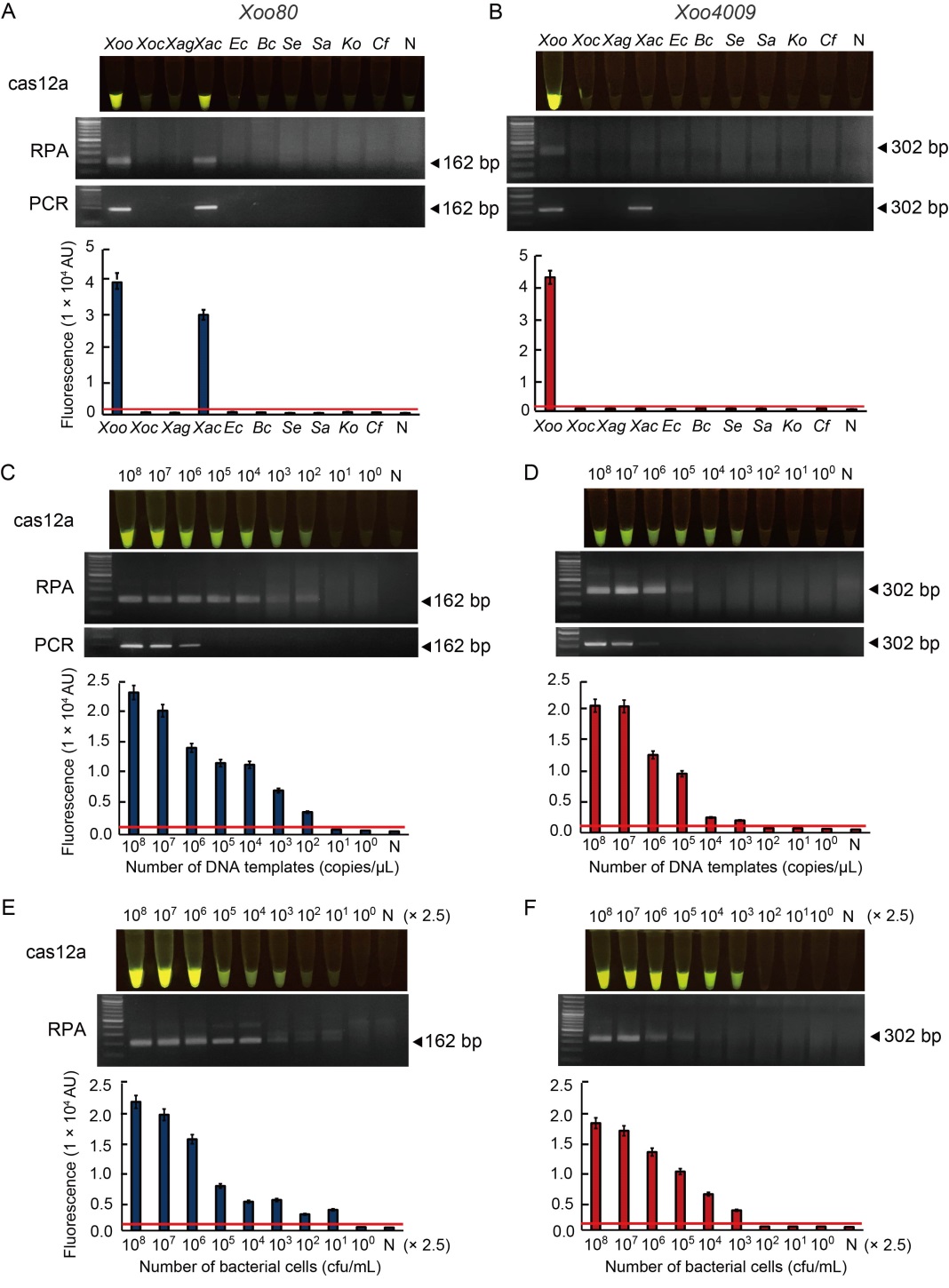
Fig. 3. Specific and sensitive assessments of RAC system for Xoo detection compared to RPA and PCR systems. A and B, Specificity test of RAC system using Xoo80 (A) and Xoo4009 loci (B) was performed with different Xanthomonas species and other bacterial species. C?F, Sensitivity test of RAC system using Xoo80 (C and E) and Xoo4009 loci (D and F) was determined with DNA templates from plasmid DNA (pUC57-Xoo80 or pUC57-Xoo4009) ranging from 0 to 1 × 108 copies/µL (C and D), and bacterial number ranged in 0?2.5 × 108 cfu/mL (E and F). RPA, Recombinase polymerase amplification; RAC, RPA-assisted cas12a. N refers to an RPA reaction without DNA templates as a negative control.
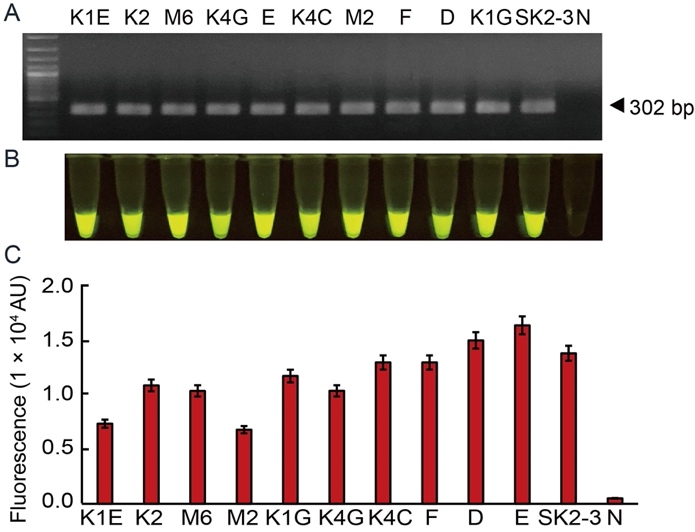
Fig. 4. Compatibility test of optimal RAC system using Xoo4009 locus. Detection feasibility of RAC used 11 Xoo isolates (K1E, K2, M6, K4G, E, K4C, M2, F, D, K1G and SK2-3) distributed in the endemic area. The resulting products were visualized by agarose gel electrophoresis (A) and an LED transilluminator (B), and the fluorescence measurement was quantified using a real-time machine (C). N refers to an RPA reaction without DNA templates as a negative control. RPA, Recombinase polymerase amplification; RAC, RPA-assisted cas12a.
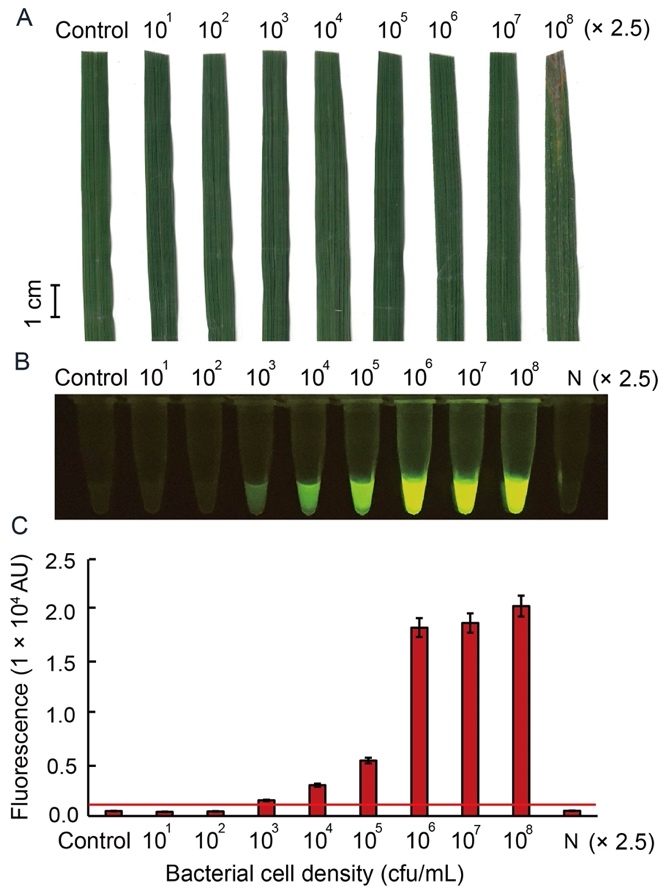
Fig. 5. Sensitivity of RAC system for Xoo detection in artificially infected rice leaf, RD47 variety, with different bacterial cell intensities. Sixty-day-old rice leaves were inoculated with XooD at different concentrations ranging from 25 to 2.5 × 108 cfu/mL for 7 d post- inoculation. External bacterial leaf blight symptoms were photographed (A). Leaf extracts were used as DNA templates in an in vitro RAC system and fluorescent signals from the RAC products were visualized by an LED transilluminator (B), and quantified by a real-time PCR machine (C). N refers to an RPA reaction without DNA templates as a negative control. RPA, Recombinase polymerase amplification; RAC, RPA-assisted cas12a.
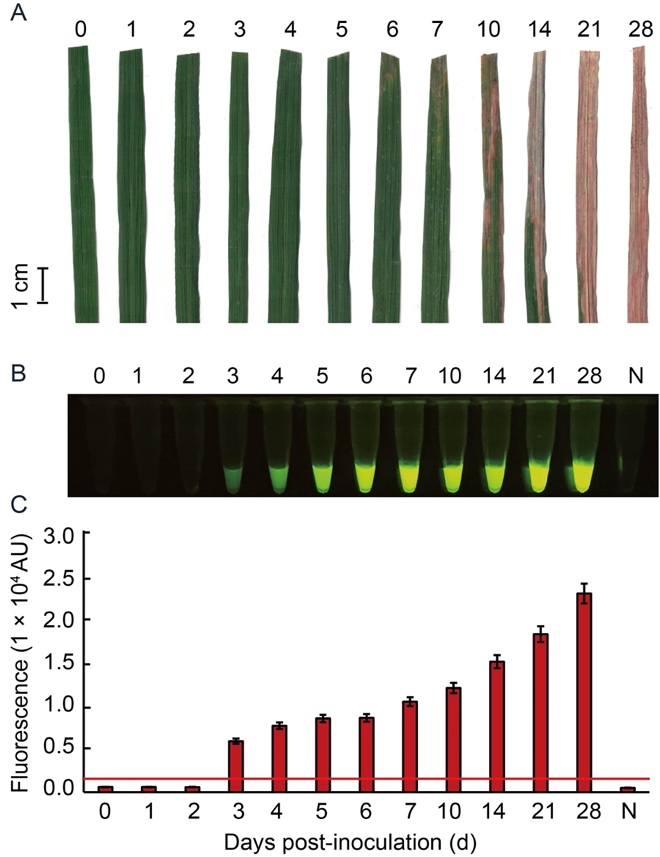
Fig. 6. Sensitivity of RAC system for Xoo detection at different days post-inoculation (dpi) of rice leaves. Leaf samples of rice variety RD47 were artificially inoculated with XooD at 2.5 × 108 cfu/mL by the clipping method. Leaves were collected at different time courses of 1-28 dpi for Xoo detection. External bacterial leaf blight symptoms were photographed (A). Leaf extracts were used as DNA templates in an in vitro RAC system and fluorescent signals from the RAC products were visualized by an LED transilluminator (B), and quantified by a real-time PCR machine (C). N refers to an RPA reaction without DNA templates as a negative control. RPA, Recombinase polymerase amplification; RAC, RPA-assisted cas12a.
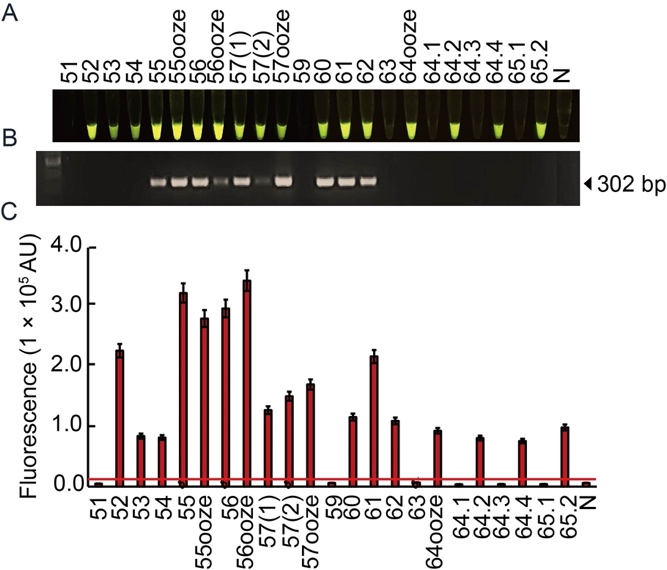
Fig. 7. Diagnosis of Xoo pathogen using RAC system in naturally infected rice samples. A total of 23 leaf samples were collected from rice fields in lower northern of Thailand. Plant extracts were used for DNA templates in the RAC and PCR reactions. The cas12a-mediated trans-cleavage fluorescence signals from the RAC assay were detected under an LED transilluminator (A), PCR products were visualized by agarose gel electrophoresis (B), and their intensities were quantified by a real-time PCR machine (C). N refers to an RPA reaction without DNA templates as a negative control. RPA, Recombinase polymerase amplification; RAC, RPA-assisted cas12a.
| [1] | Adli M. 2018. The CRISPR tool kit for genome editing and beyond. Nat Commun, 9(1): 1911. |
| [2] | Ai J W, Zhou X, Xu T, Yang M L, Chen Y Y, He G Q, Pan N, Cai Y W, Li Y J, Wang X R, Su H, Wang T, Zeng W Q, Zhang W H. 2019. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg Microbes Infect, 8(1): 1361-1369. |
| [3] | Ali Z, Aman R, Mahas A, Rao G S, Tehseen M, Marsic T, Salunke R, Subudhi A K, Hala S M, Hamdan S M, Pain A, Alofi F S, Alsomali A, Hashem A M, Khogeer A, Almontashiri N A M, Abedalthagafi M, Hassan N, Mahfouz M M. 2020. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res, 288: 198129. |
| [4] | Aman R, Mahas A, Marsic T, Hassan N, Mahfouz M M. 2020. Efficient, rapid, and sensitive detection of plant RNA viruses with one-pot RT-RPA-CRISPR/Cas12a assay. Front Microbiol, 11: 610872. |
| [5] | Ansari T H, Haque S S, Uddin A. 2020. Pest management decision guide: Green and yellow list: Management of bacterial leaf blight (BLB) of rice. Plantwise knowledge Bank, CAB International. [2 October, 2021]. https://www.plantwise.org/KnowledgeBank/pmdg/20167800391. |
| [6] | Bai J, Lin H S, Li H J, Zhou Y, Liu J S, Zhong G R, Wu L T, Jiang W F, Du H L, Yang J Y, Xie Q M, Huang L Z. 2019. Cas12a-based on-site and rapid nucleic acid detection of African swine fever. Front Microbiol, 10: 2830. |
| [7] | Barra G B, Santa Rita T H, de Almeida Vasques J, Chianca C F, Nery L F A, Santana Soares Costa S. 2015. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem, 48(15): 976-981. |
| [8] |
Barrangou R, Horvath P. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol, 2: 17092.
PMID |
| [9] | Buddhachat K, Ritbamrung O, Sripairoj N, Inthima P, Ratanasut K, Boonsrangsom T, Sujipuli K. 2021. One-step colorimetric LAMP (cLAMP) assay for visual detection of Xanthomonas oryzae pv. oryzae in rice. Crop Prot, 150: 105809. |
| [10] |
Carpenter S C D, Mishra P, Ghoshal C, Dash P K, Wang L, Midha S, Laha G S, Lore J S, Kositratana W, Singh N K, Singh K, Patil P B, Oliva R, Patarapuwadol S, Bogdanove A J, Rai R. 2018. A strain of an emerging Indian Xanthomonas oryzae pv. oryzae pathotype defeats the rice bacterial blight resistance gene xa13 without inducing a clade III SWEET gene and is nearly identical to a recent Thai isolate. Front Microbiol, 9: 2703.
PMID |
| [11] | Chaijarasphong T, Thammachai T, Itsathitphaisarn O, Sritunyalucksana K, Suebsing R. 2019. Potential application of CRISPR-Cas12a fluorescence assay coupled with rapid nucleic acid amplification for detection of white spot syndrome virus in shrimp. Aquaculture, 512: 734340. |
| [12] | Chen J S, Ma E B, Harrington L B, da Costa M, Tian X R, Palefsky J M, Doudna J A. 2018. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360: 436-439. |
| [13] | Chen P, Zhou J, Wan Y B, Liu H, Li Y Z, Liu Z X, Wang H J, Lei J, Zhao K, Zhang Y L, Wang Y, Zhang X H, Yin L. 2020. A Cas12a ortholog with stringent PAM recognition followed by low off-target editing rates for genome editing. Genome Biol, 21(1): 78. |
| [14] |
Creutzburg S C A, Swartjes T, van der Oost J. 2020. Medium- throughput in vitro detection of DNA cleavage by CRISPR- Cas12a. Methods, 172: 27-31.
PMID |
| [15] |
Cui Z, Ojaghian M R, Tao Z, Kakar K U, Zeng J, Zhao W, Duan Y, Vera Cruz C M, Li B, Zhu B, Xie G. 2016. Multiplex PCR assay for simultaneous detection of six major bacterial pathogens of rice. J Appl Microbiol, 120(5): 1357-1367.
PMID |
| [16] |
Daher R K, Stewart G, Boissinot M, Bergeron M G. 2016. Recombinase polymerase amplification for diagnostic applications. Clin Chem, 62(7): 947-958.
PMID |
| [17] | Ding X, Yin K, Li Z Y, Lalla R V, Ballesteros E, Sfeir M M, Liu C C. 2020. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun, 11(1): 4711. |
| [18] |
Garita-Cambronero J, Palacio-Bielsa A, López M M, Cubero J. 2017. Pan-genomic analysis permits differentiation of virulent and non-virulent strains of Xanthomonas arboricola that cohabit Prunus spp. and elucidate bacterial virulence factors. Front Microbiol, 8: 573.
PMID |
| [19] |
Gootenberg J S, Abudayyeh O O, Lee J W, Essletzbichler P, Dy A J, Joung J, Verdine V, Donghia N, Daringer N M, Freije C A, Myhrvold C, Bhattacharyya R P, Livny J, Regev A, Koonin E V, Hung D T, Sabeti P C, Collins J J, Zhang F. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356: 438-442.
PMID |
| [20] | Kanitchinda S, Srisala J, Suebsing R, Prachumwat A, Chaijarasphong T. 2020. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol Rep, 27: e00485. |
| [21] | Lang J M, Hamilton J P, Diaz M G Q, van Sluys M A, Burgos M R G, Vera Cruz C M, Buell C R, Tisserat N A, Leach J E. 2010. Genomics-based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis, 94(3): 311-319. |
| [22] | Lang J M, Langlois P, Nguyen M H R, Triplett L R, Purdie L, Holton T A, Djikeng A, Vera Cruz C M, Verdier V, Leach J E. 2014. Sensitive detection of Xanthomonas oryzae Pathovars oryzae and oryzicola by loop-mediated isothermal amplification. Appl Environ Microbiol, 80(15): 4519-4530. |
| [23] | Lau H Y, Botella J R. 2017. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front Plant Sci, 8: 2016. |
| [24] | le Thanh T, Thumanu K, Wongkaew S, Boonkerd N, Teaumroong N, Phansak P, Buensanteai N. 2017. Salicylic acid-induced accumulation of biochemical components associated with resistance against Xanthomonas oryzae pv. oryzae in rice. J Plant Interact, 12(1): 108-120. |
| [25] | Li S Y, Cheng Q X, Wang J M, Li X Y, Zhang Z L, Gao S, Cao R B, Zhao G P, Wang J. 2018a. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov, 4: 20. |
| [26] | Li S Y, Cheng Q X, Liu J K, Nie X Q, Zhao G P, Wang J. 2018b. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res, 28(4): 491-493. |
| [27] | Liu H, Wang J B, Zeng H J, Liu X F, Jiang W, Wang Y, Ouyang W B, Tang X M. 2021. RPA-Cas12a-FS: A frontline nucleic acid rapid detection system for food safety based on CRISPR- Cas12a combined with recombinase polymerase amplification. Food Chem, 334: 127608. |
| [28] | Liu P P, Luk K, Shin M, Idrizi F, Kwok S, Roscoe B, Mintzer E, Suresh S, Morrison K, Frazão J B, Bolukbasi M F, Ponnienselvan K, Luban J, Zhu L J, Lawson N D, Wolfe S A. 2019. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res, 47(8): 4169-4180. |
| [29] | Lu W, Pan L Q, Zhao H J, Jia Y L, Wang Y L, Yu X P, Wang X Y. 2014. Molecular detection of Xanthomonas oryzae pv. oryzae, Xanthomonas oryzae pv. oryzicola, and Burkholderia glumae in infected rice seeds and leaves. Crop J, 2(6): 398-406. |
| [30] | Mahas A, Hassan N, Aman R, Marsic T, Wang Q C, Ali Z, Mahfouz M M. 2021. LAMP-coupled CRISPR-Cas12a module for rapid and sensitive detection of plant DNA viruses. Viruses, 13(3): 466. |
| [31] |
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res, 28: E63.
PMID |
| [32] | Piepenburg O, Williams C H, Stemple D L, Armes N A. 2006. DNA detection using recombination proteins. PLoS Biol, 4(7): e204. |
| [33] | Rudresha G V, Urs A P, Manjuprasanna V N, Suvilesh K N, Sharanappa P, Vishwanath B S. 2019. Plant DNases are potent therapeutic agents against Echis carinatus venom-induced tissue necrosis in mice. J Cell Biochem, 120(5): 8319-8332. |
| [34] |
Sullivan T J, Dhar A K, Cruz-Flores R, Bodnar A G. 2019. Rapid, CRISPR-based, field-deployable detection of white spot syndrome virus in shrimp. Sci Rep, 9(1): 19702.
PMID |
| [35] | Wang X J, Ji P P, Fan H Y, Dang L, Wan W W, Liu S Y, Li Y H, Yu W X, Li X Y, Ma X D, Ma X, Zhao Q, Huang X X, Liao M. 2020. CRISPR/Cas12a technology combined with immune- chromatographic strips for portable detection of African swine fever virus. Commun Biol, 3(1): 62. |
| [36] | Xiao G H, Zhang S, Liang Z H, Li G Q, Fang M T, Liu Y Y, Zhang J J, Ou M, He X, Zhang T Y, Zeng C C, Liu L, Zhang G L. 2020. Identification of Mycobacterium abscessus species and subspecies using the Cas12a/sgRNA-based nucleic acid detection platform. Eur J Clin Microbiol Infect Dis, 39(3): 551-558. |
| [37] | Xu H P, Zhang X L, Cai Z X, Dong X Q, Chen G, Li Z L, Qiu L M, He L, Liang B, Liu X L, Liu J F. 2020. An isothermal method for sensitive detection of Mycobacterium tuberculosis complex using clustered regularly interspaced short palindromic repeats/ Cas12a cis and trans cleavage. J Mol Diagn, 22(8): 1020-1029. |
| [38] | Xu L Q, Dai Q Q, Shi Z Y, Liu X T, Gao L, Wang Z Z, Zhu X Y, Li Z. 2020. Accurate MRSA identification through dual- functional aptamer and CRISPR-Cas12a assisted rolling circle amplification. J Microbiol Methods, 173: 105917. |
| [39] | Yasmin S, Hafeez F Y, Mirza M S, Rasul M, Arshad H M I, Zubair M, Iqbal M. 2017. Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front Microbiol, 8: 1895. |
| [40] | Yuan M, Ke Y G, Huang R Y, Ma L, Yang Z Y, Chu Z H, Xiao J H, Li X H, Wang S P. 2016. A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife, 5: e19605. |
| [41] |
Zetsche B, Gootenberg J S, Abudayyeh O O, Slaymaker I M, Makarova K S, Essletzbichler P, Volz S E, Joung J, van der Oost J, Regev A, Koonin E V, Zhang F. 2015. Cpf1 is a single RNA- guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3): 759-771.
PMID |
| [42] | Zhang Y M, Zhang Y, Xie K. 2020. Evaluation of CRISPR/ Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice. Mol Breed, 40(1): 1-12. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||