
Rice Science ›› 2017, Vol. 24 ›› Issue (5): 291-298.DOI: 10.1016/j.rsci.2017.04.005
• Orginal Article • Previous Articles
Saakre Manjesh( ), Meera Baburao Thirthikar, Puthenpeedikal Salim Abida, Mary Ffancies Rose, Poothecty Achuthan Valasala, Thomas George, Radha Sivarajan Sajeevan
), Meera Baburao Thirthikar, Puthenpeedikal Salim Abida, Mary Ffancies Rose, Poothecty Achuthan Valasala, Thomas George, Radha Sivarajan Sajeevan
Received:2016-12-21
Accepted:2017-04-20
Online:2017-09-15
Published:2017-08-31
Saakre Manjesh, Meera Baburao Thirthikar, Puthenpeedikal Salim Abida, Mary Ffancies Rose, Poothecty Achuthan Valasala, Thomas George, Radha Sivarajan Sajeevan. Identification and Characterization of Genes Responsible for Drought Tolerance in Rice Mediated by Pseudomonas fluorescens[J]. Rice Science, 2017, 24(5): 291-298.
Add to citation manager EndNote|Ris|BibTeX
| Gene | Sequence (5′-3′) | Fragment length (bp) |
|---|---|---|
| COX1 | F: CTCCTAGTCGGCCTGATTTC | 108 |
| R: CATGAGCAGTAGCATCCTTGA | ||
| PKDP | F: CGTTGATAGTCGCCGCTAAA | 109 |
| R: TTTAAGAGGCGGGAATGGTG | ||
| bZIP1 | F: GAGCGTACTCTGTCCCATTTAG | 115 |
| R: GTTCCAGCGATGAGGTTGT | ||
| AP2- EREBP | F: AGGTAAAGCCCGAGCAATTC | 101 |
| R: GCATCGGTGAATGGTGGTATAA | ||
| Hsp20 | F: TGTGTGTCACCACGCTTTA | 119 |
| R: CCTCGCATAGACCCATTCATC | ||
| COC1 | F: CACCTCATGACGATGCAAGA | 101 |
| R: GAGCTTGCTCACTCCTTCAA |
Table 1 Primers used by real-time PCR.
| Gene | Sequence (5′-3′) | Fragment length (bp) |
|---|---|---|
| COX1 | F: CTCCTAGTCGGCCTGATTTC | 108 |
| R: CATGAGCAGTAGCATCCTTGA | ||
| PKDP | F: CGTTGATAGTCGCCGCTAAA | 109 |
| R: TTTAAGAGGCGGGAATGGTG | ||
| bZIP1 | F: GAGCGTACTCTGTCCCATTTAG | 115 |
| R: GTTCCAGCGATGAGGTTGT | ||
| AP2- EREBP | F: AGGTAAAGCCCGAGCAATTC | 101 |
| R: GCATCGGTGAATGGTGGTATAA | ||
| Hsp20 | F: TGTGTGTCACCACGCTTTA | 119 |
| R: CCTCGCATAGACCCATTCATC | ||
| COC1 | F: CACCTCATGACGATGCAAGA | 101 |
| R: GAGCTTGCTCACTCCTTCAA |
| Treatment | Shoot length (cm) | Root length (cm) | Fresh weight (g) | Dry weight (g) | Yield per panicle (g) | 1000-grain weight (g) |
|---|---|---|---|---|---|---|
| Control | 111.50 | 26.66 | 45.620 | 29.500 | 3.196 | 24.520 |
| Water stressed | 91.38 | 15.54 | 27.420 | 11.800 | 1.482 | 18.700 |
| Water stressed + Pf1-treated | 98.86 | 24.00 | 34.940 | 14.400 | 1.994 | 21.220 |
| CD (1%) | 11.59 | 3.04 | 10.248 | 12.584 | 0.297 | 1.747 |
Table 2 Measurement of biometric parameters after harvest using Duncan’s multiple range statistical analysis (n = 3).
| Treatment | Shoot length (cm) | Root length (cm) | Fresh weight (g) | Dry weight (g) | Yield per panicle (g) | 1000-grain weight (g) |
|---|---|---|---|---|---|---|
| Control | 111.50 | 26.66 | 45.620 | 29.500 | 3.196 | 24.520 |
| Water stressed | 91.38 | 15.54 | 27.420 | 11.800 | 1.482 | 18.700 |
| Water stressed + Pf1-treated | 98.86 | 24.00 | 34.940 | 14.400 | 1.994 | 21.220 |
| CD (1%) | 11.59 | 3.04 | 10.248 | 12.584 | 0.297 | 1.747 |
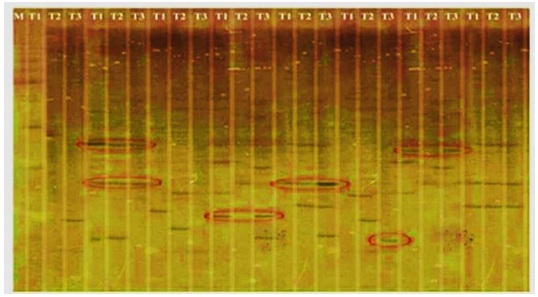
Fig. 1. Differential display pattern of transcript derived fragments from three treatments in rice variety PTB45.^M, Marker; T1, Control; T2, Water stressed; T3, Water stressed + Pf1-treated.
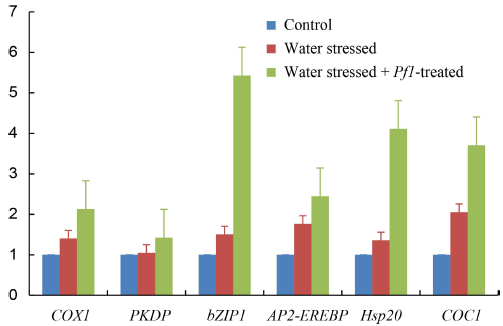
Fig. 2. qRT-PCR analysis of COX1, PKDP, bZIP1, AP2-EREBP, Hsp20 and COC1 genes under three different treatments in 65 d old plants of rice variety PTB45. ^Actin was used as endogenous control. Error bars represent standard deviation.
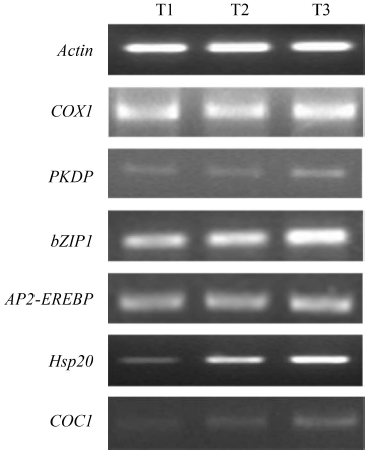
Fig. 3. Semi-quantitative reverse transcriptase PCR analysis showing differential gene expressions in different treatments.^T1, Control; T2, Water stressed; T3, Water stressed + Pf1-treated.
| Sequence | Gene code | Gene description | Length (bp) | Hit score | E-value | Accession ID | Database |
| Sequence 1 | COX1 | Cytochrome c oxidase subunit 1 | 159 | 150 | 0.00 | Os12g0561000 | IGR, rice |
| Sequence 2 | PKDP | Protein kinase domain protein | 161 | 129 | 0.00 | Os08g39170 | IGR, rice |
| Sequence 3 | bZIP1 | bZIP family protein | 680 | 83 | 2e-17 | CT833525 | PTFD |
| Sequence 4 | AP2-EREBP | APETALA2-ethylene responsive element binding protein | 276 | 79 | 0.00 | OsIBCD031146 | DRTF |
| Sequence 5 | Hsp20 | Heat shock protein 20 | 484 | 230 | 4e-25 | Os01g04380.1 | RGAP |
| Sequence 6 | COC1 | Circadian oscillator component 1 | 228 | 97 | 3e-06 | AY885936 | PTFD |
| IGR, Institute of Genome Research; PTFD, Plant Transcription Factor Database; DRTF, Database of Rice Transcription Factor; RGAP, Rice Genome Annotation Project. | |||||||
Table 3 Sequence homologies of the TDFs with known genes based on BLAST from different databases.
| Sequence | Gene code | Gene description | Length (bp) | Hit score | E-value | Accession ID | Database |
| Sequence 1 | COX1 | Cytochrome c oxidase subunit 1 | 159 | 150 | 0.00 | Os12g0561000 | IGR, rice |
| Sequence 2 | PKDP | Protein kinase domain protein | 161 | 129 | 0.00 | Os08g39170 | IGR, rice |
| Sequence 3 | bZIP1 | bZIP family protein | 680 | 83 | 2e-17 | CT833525 | PTFD |
| Sequence 4 | AP2-EREBP | APETALA2-ethylene responsive element binding protein | 276 | 79 | 0.00 | OsIBCD031146 | DRTF |
| Sequence 5 | Hsp20 | Heat shock protein 20 | 484 | 230 | 4e-25 | Os01g04380.1 | RGAP |
| Sequence 6 | COC1 | Circadian oscillator component 1 | 228 | 97 | 3e-06 | AY885936 | PTFD |
| IGR, Institute of Genome Research; PTFD, Plant Transcription Factor Database; DRTF, Database of Rice Transcription Factor; RGAP, Rice Genome Annotation Project. | |||||||
| [1] | Auffhammer M, Ramanathan V, Vincent J R.2011. Climate change, the monsoon, and rice yield in India.Clim Change, 111(2): 411-424. |
| [2] | Ahmad M, Kibert M.2013. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud Univ Sci, 26(1): 1-20. |
| [3] | Arshad M, Saleem M, Hussain S.2007. Perspectives of bacterial ACC deaminase in phytoremediation.Trends Biotechnol, 25(8): 356-362. |
| [4] | Bononi A, Agnoletto C, de Marchi E, Marchi S, Patergnani S, Bonora M, Giorgi C, Missiroli S, Poletti F, Rimessi A, Pinton P. 2011. Protein kinases and phosphatases in the control of cell fate.Enzyme Res, 2011: 1-26. |
| [5] | Chen L H, Han J P, Deng X M, Tan S L, Li L L, Li L, Zhou J F, Peng H, Yang G X, He G Y, Zhang W X.2016. Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon.Sci Rep, 6: 21623. |
| [6] | Deveau A, Palin B, Delaruelle C, Peter M, Kohler A, Pierrat J C, Pierrart J C, Sarniguet A, Garbaye J, Martin F, Frey-Klett P.2007. The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N.New Phytol, 175(4): 743-755. |
| [7] | Dominguez-Nunez J A, Munoz D, de la Cruz A, Saiz de Omenaca J A.2013. Effects of Pseudomonas fluorescens on the water parameters of mycorrhizal and non-mycorrhizal seedlings of Pinus halepensis.Agron, 3(3): 571-582. |
| [8] | Ganeshan G, Arthikala A M.2005. Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases.J Plant Interact, 1(3): 123-134. |
| [9] | Ghadirnezhad R, Fallah A.2014. Temperature effect on yield and yield components of different rice cultivars in flowering stage.Int J Agron, 2014: 4. |
| [10] | Halliwell B.2006. Reactive species and antioxidants: Redox biology is a fundamental theme of aerobic life.Plant Physiol, 141(2): 312-322. |
| [11] | Heinonsalo J, Hurme K R, Sen R.2004. Recent 14C-labelled assimilate allocation to Scots pine seedling root and mycorrhizosphere compartments developed on reconstructed podsol humus, E- and B-mineral horizons.Plant Soil, 259: 111-121. |
| [12] | Herman M A B, Nault B A, Smart C D.2008. Effects of plant growth promoting rhizobacteria on bell pepper production and green peach aphid infestations in New York.Crop Prot, 27(6): 996-1002. |
| [13] | IRRI (International Rice Research Institute). 1996. Standard Evaluation System for Rice. Los Banos, the Philippines: International Rice Research Institute: 102. |
| [14] | Lugtenbergc B, Kamilova F.2009. Plant-growth-promoting rhizobacteria.Annu Rev Microbiol, 63: 541-556. |
| [15] | Matthijs S, Tehrani K A, Laus G, Jackson R W, Cooper R W, Cornelis P.2007. Thioquinolobactin, a Pseudomonas siderophore with antifungal and anti-pythium activity.Environ Microbiol, 9(2): 425-434. |
| [16] | Riechmann J L, Meyerowitz E M.1998. The AP2/EREBP family of plant transcription factors.Biol Chem, 379(6): 633-646. |
| [17] | Scheeff E D, Bourne P E.2005. Structural evolution of the protein kinase-like superfamily.PLoS Comput Biol, 1(5): 351-381. |
| [18] | Singh K B, Foley R C, Onate-Sanchez L.2002. Transcription factors in plant defense and stress responses.Curr Opin Plant Biol, 5(5): 430-436. |
| [19] | Vleesschauwer D D, Djavaheri M, Bakker P A H M, Hofte M.2008. Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response.Plant Physiol, 148: 1996-2012. |
| [20] | Yan S P, Tang Z C, Su W A, Sun W N.2005. Proteomic analysis of salt stress responsive proteins in rice root.Proteomics, 5(1): 235-244. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [13] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||