
Rice Science ›› 2019, Vol. 26 ›› Issue (3): 147-156.DOI: 10.1016/j.rsci.2018.08.006
• Research Papers • Previous Articles Next Articles
Khlaimongkhon Sudthana1, Chakhonkaen Sriprapai2, Pitngam Keasinee2, Ditthab Khanittha2, Sangarwut Numphet2, Panyawut Natjaree2, Wasinanon Thiwawan2, Mongkolsiriwatana Chareerat3, Chunwongse Julapark1, Muangprom Amorntip1,2( )
)
Received:2018-07-09
Accepted:2018-08-24
Online:2019-05-28
Published:2019-01-25
Khlaimongkhon Sudthana, Chakhonkaen Sriprapai, Pitngam Keasinee, Ditthab Khanittha, Sangarwut Numphet, Panyawut Natjaree, Wasinanon Thiwawan, Mongkolsiriwatana Chareerat, Chunwongse Julapark, Muangprom Amorntip. Molecular Markers and Candidate Genes for Thermo-Sensitive Genic Male Sterile in Rice[J]. Rice Science, 2019, 26(3): 147-156.
Add to citation manager EndNote|Ris|BibTeX

Fig. 1. Pollen staining of thermo-sensitive genic male sterility (TGMS) mutant and wild type. A, Abortive anther of the TGMS mutant grown under high temperature (32 ± 2) ºC. B, Normal anther of the TGMS mutant grown under low temperature (24 ± 2) ºC. C, Normal anther of wild type grown under high temperature (32 ± 2) ºC. D, Normal anther of wild type grown under low temperature (24 ± 2) ºC.
| F2 population | Total No. of lines | No. of fertile individuals | No. of sterile individuals | Segregation rate | χ2 |
|---|---|---|---|---|---|
| B2 × B30 | 917 | 700 | 217 | 3.22 | 0.91 |
| B2 × B11 | 981 | 722 | 259 | 2.79 | 0.98 |
| B2 × KM9 | 1 784 | 1 334 | 450 | 2.96 | 0.042 |
Table 1 Segregation patterns of F2 populations for fertility and sterility.
| F2 population | Total No. of lines | No. of fertile individuals | No. of sterile individuals | Segregation rate | χ2 |
|---|---|---|---|---|---|
| B2 × B30 | 917 | 700 | 217 | 3.22 | 0.91 |
| B2 × B11 | 981 | 722 | 259 | 2.79 | 0.98 |
| B2 × KM9 | 1 784 | 1 334 | 450 | 2.96 | 0.042 |
| Marker | Forward primers (5´-3´) | Reverse primers (5´-3´) | Position (Mb) |
|---|---|---|---|
| RM12557 | AGCACCACCTCCTCGAACTCC | CAACCCTACCTTGCTTCTTCTTGC | 4.19 |
| RM4355 | GGGATGAGAGTAGAAGGCACAAGG | GCTTAATGCCTTTGATCGTTGC | 4.26 |
| RM12593 | ACAATACGCTGTGCCAATCTGC | TATTGGCCGTGATGATGAAGTGC | 4.57 |
| RM12601 | GCGAACTCGACGACTACTCAACC | CACAACGTCGTCTCCAAGTGC | 4.69 |
| RM7638 | GGCTGTCCGTCTTGTAGTGAGAAGC | TCCTTGCAACTTCCGGAGACC | 4.86 |
| RM12632 | GGCTTTATTTGTTCTGGCCTTGTGG | CCAAATCAAATGGGTCCTTTGTGC | 5.08 |
| RM12649 | TTGAGGTAGGGCACGAGGTCTGG | AATCCGAATGGGACCGAATCACC | 5.25 |
| RM12655 | AGGACCAATCCAAAGCGTTTAGC | CCTGCAGTAGATTGCATTGAACC | 5.34 |
| RM5664 | GTTCGGCTCCACCTAAACCAAGC | GGCATTCGTCTCGTCTTTGAGG | 5.37 |
| RM6378 | CTGATCATCTCATGCCTCCTACG | TCCATCTCCCAATATGACCAACC | 5.47 |
| 2gAP004869 | AGCTTAAACATATTGGCTGCACG | TAGCCAAGCACGTATACACTCCC | 5.67 |
| RM12674 | TAAATGCCAACCAACTCCAAGC | AACTGCGTTTGGGAATATCTCG | 5.68 |
| RM12676 | ACTGACGATTGGGCACATTATTCC | CTGCAAATTGGTGGGTGATTGC | 5.74 |
| 2gAP004045 | CCGAAACCATATATCCAACACGA | TCAAATGTTATTTGGGCGTAACTTT | 5.76 |
| 2gAP0050058 | CAAACAACCACTTTTGTTCAGCA | ATATATGCTGTGGGCAGGATCAA | 5.81 |
| 2gAP003974 | CAATTCGGAGATTACATTCGGTC | ACTATAGACGCAGCTTGTGTGCC | 6 .0 |
| vf0206114052 | CGAGACCAATCGATCCTCCA | TAACCTGCATCTGTGGTGGA | 6.11 |
| 2gAP004085 | TGGCAACTAATAGGCATCGTCG | TCTTGTAGCTGCAACATCCCCC | 6.26 |
| 2gAP004086 | TCAAGCAATCACAACCCCTTCC | GGCTACCCAAACCATGCATCA | 6.28 |
| Os02g12300 | TCCCATTTAATTTGGTTCATCG | GCTAACGTGCGTGGATTGAG | 6.4 |
| Os02g12350 | TCCATCTGCAAATCCATAGCA | TCTGGTGCATAGCTGCTGGT | 6.42 |
| Os02g12370 | ACTTATGCGGTGGTCAAGCA | TTGTTCCTCCACGGCTGATA | 6.44 |
| 2gAP005394 | TGCAAACAGTTGGATTAGCTTGA | TGACAGTGCGCTTATCTTCCATA | 7.17 |
| 2gAP005756 | AATTCATTGGACGCACACACAT | TGCATTGGGAAATTGAAAAGATT | 7.26 |
| 2gAP004070 | ATGGTGCCGGATAACGTATTACA | GGCTTGCGGTAAGAATATAGGAGA | 7.44 |
Supplemental Table 1 Markers on chromosome 2 linked to the tms gene using F2 male sterile plants from B2 × B30, B2 × B11 and B2 × KM9.
| Marker | Forward primers (5´-3´) | Reverse primers (5´-3´) | Position (Mb) |
|---|---|---|---|
| RM12557 | AGCACCACCTCCTCGAACTCC | CAACCCTACCTTGCTTCTTCTTGC | 4.19 |
| RM4355 | GGGATGAGAGTAGAAGGCACAAGG | GCTTAATGCCTTTGATCGTTGC | 4.26 |
| RM12593 | ACAATACGCTGTGCCAATCTGC | TATTGGCCGTGATGATGAAGTGC | 4.57 |
| RM12601 | GCGAACTCGACGACTACTCAACC | CACAACGTCGTCTCCAAGTGC | 4.69 |
| RM7638 | GGCTGTCCGTCTTGTAGTGAGAAGC | TCCTTGCAACTTCCGGAGACC | 4.86 |
| RM12632 | GGCTTTATTTGTTCTGGCCTTGTGG | CCAAATCAAATGGGTCCTTTGTGC | 5.08 |
| RM12649 | TTGAGGTAGGGCACGAGGTCTGG | AATCCGAATGGGACCGAATCACC | 5.25 |
| RM12655 | AGGACCAATCCAAAGCGTTTAGC | CCTGCAGTAGATTGCATTGAACC | 5.34 |
| RM5664 | GTTCGGCTCCACCTAAACCAAGC | GGCATTCGTCTCGTCTTTGAGG | 5.37 |
| RM6378 | CTGATCATCTCATGCCTCCTACG | TCCATCTCCCAATATGACCAACC | 5.47 |
| 2gAP004869 | AGCTTAAACATATTGGCTGCACG | TAGCCAAGCACGTATACACTCCC | 5.67 |
| RM12674 | TAAATGCCAACCAACTCCAAGC | AACTGCGTTTGGGAATATCTCG | 5.68 |
| RM12676 | ACTGACGATTGGGCACATTATTCC | CTGCAAATTGGTGGGTGATTGC | 5.74 |
| 2gAP004045 | CCGAAACCATATATCCAACACGA | TCAAATGTTATTTGGGCGTAACTTT | 5.76 |
| 2gAP0050058 | CAAACAACCACTTTTGTTCAGCA | ATATATGCTGTGGGCAGGATCAA | 5.81 |
| 2gAP003974 | CAATTCGGAGATTACATTCGGTC | ACTATAGACGCAGCTTGTGTGCC | 6 .0 |
| vf0206114052 | CGAGACCAATCGATCCTCCA | TAACCTGCATCTGTGGTGGA | 6.11 |
| 2gAP004085 | TGGCAACTAATAGGCATCGTCG | TCTTGTAGCTGCAACATCCCCC | 6.26 |
| 2gAP004086 | TCAAGCAATCACAACCCCTTCC | GGCTACCCAAACCATGCATCA | 6.28 |
| Os02g12300 | TCCCATTTAATTTGGTTCATCG | GCTAACGTGCGTGGATTGAG | 6.4 |
| Os02g12350 | TCCATCTGCAAATCCATAGCA | TCTGGTGCATAGCTGCTGGT | 6.42 |
| Os02g12370 | ACTTATGCGGTGGTCAAGCA | TTGTTCCTCCACGGCTGATA | 6.44 |
| 2gAP005394 | TGCAAACAGTTGGATTAGCTTGA | TGACAGTGCGCTTATCTTCCATA | 7.17 |
| 2gAP005756 | AATTCATTGGACGCACACACAT | TGCATTGGGAAATTGAAAAGATT | 7.26 |
| 2gAP004070 | ATGGTGCCGGATAACGTATTACA | GGCTTGCGGTAAGAATATAGGAGA | 7.44 |
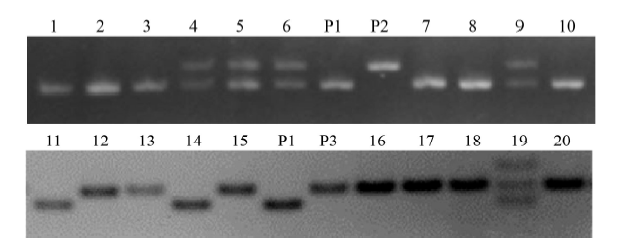
Fig. 2. Samples of genotyping F2 male sterile plants. P1, Female parent (IR68301S, B2); P2, Male parent (IR14632, B30); 1-10, F2 male sterile individuals of B2 × B30 (indica × japonica) using Os02g12370 marker; P3, Male parent (Supanburi 91062, B11); 11-20, F2 male sterile individuals of B2 × B11 (indica × indica) using vf0206114052 marker.
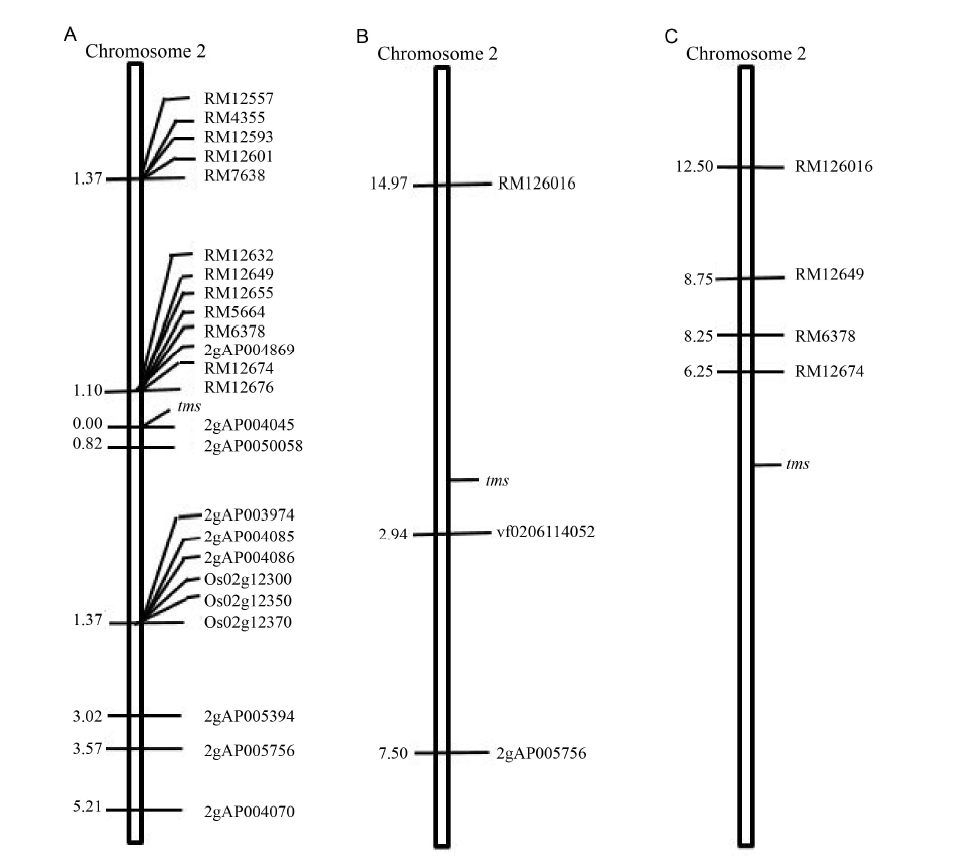
Fig. 3. Genetic linkage map of thermo-sensitive genic male sterility gene on chromosome 2.The linkage maps were analyzed based on male sterile F2 populations of IR68301S × IR14632 (A), IR68301S × Supanburi 91062 (B) and IR68301S × IR67966-188-2-2-1 (C). Distances of each marker in centiMorgans (cM) from tms gene were given on the left side of the genetic map.
| Gene | Description |
|---|---|
| Os02g0202500 | Conserved hypothetical protein |
| Os02g0202600 | Tetratricopeptide-like helical domain containing protein |
| Os02g0202800 | Zinc ion binding protein |
| Os02g0202900 | OsFBK3-F-box domain and kelch repeat containing protein |
| Os02g0202950 | Concanavalin A-like lectin/glucanases superfamily and Legume lectin domain |
| Os02g0203000 | Carbohydrate binding, sequence-specific DNA binding, sequence-specific DNA binding transcription factor activity |
| Os02g0203300 | UDP-glucuronosyl/UDP-glucosyltransferase |
| Os02g0203401 | hypothetical gene, protein coding |
| Os02g0203500 | Putative uncharacterized protein OJ1135_F06.25; Putative uncharacterized protein P0544H11.7 |
| Os02g0203700 | Zinc finger protein; Zinc finger transcription factor ZFP30 |
Table 2 Genes located between the two flanking markers RM12676 and 2gAP0050058.
| Gene | Description |
|---|---|
| Os02g0202500 | Conserved hypothetical protein |
| Os02g0202600 | Tetratricopeptide-like helical domain containing protein |
| Os02g0202800 | Zinc ion binding protein |
| Os02g0202900 | OsFBK3-F-box domain and kelch repeat containing protein |
| Os02g0202950 | Concanavalin A-like lectin/glucanases superfamily and Legume lectin domain |
| Os02g0203000 | Carbohydrate binding, sequence-specific DNA binding, sequence-specific DNA binding transcription factor activity |
| Os02g0203300 | UDP-glucuronosyl/UDP-glucosyltransferase |
| Os02g0203401 | hypothetical gene, protein coding |
| Os02g0203500 | Putative uncharacterized protein OJ1135_F06.25; Putative uncharacterized protein P0544H11.7 |
| Os02g0203700 | Zinc finger protein; Zinc finger transcription factor ZFP30 |
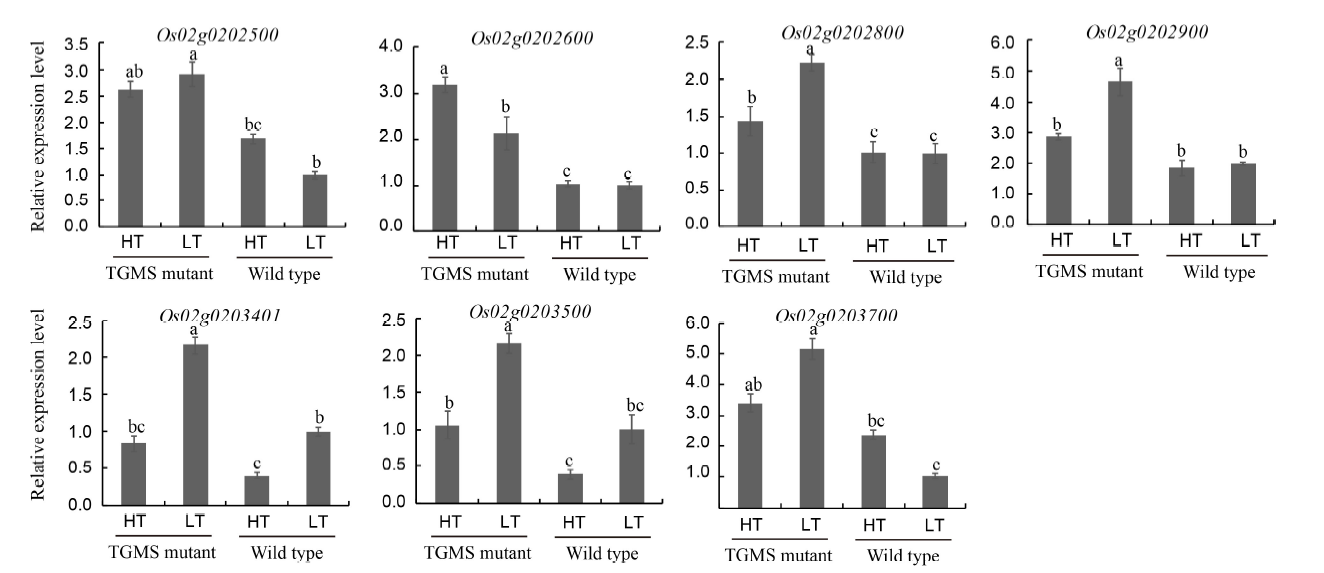
Fig. 4. Relative expression levels of genes in panicles by quantitative real-time PCR. HT, High temperature condition (32 ºC); LT, Low temperature condition (24 ºC); TGMS, Thermo-sensitive genic male sterility.Data represent Mean ± SE, and the same lowercase letter(s) indicate no significant difference at P < 0.05 by Duncan’s new multiple range test.
| Marker | Forward primers (5´-3´) | Reverse primers (5´-3´) | Position (Mb) |
|---|---|---|---|
| vf0205500813 | CGGGTCGACTTGCCTGAATA | AGAAATGGCGTTGGAAGCAG | 5.5 |
| vf0205520833 | GTCACGCCGATATCGTCAAC | ACGTCATCATCCACTAACAAGT | 5.52 |
| vf0205535398 | ACCATAATGATTCCGGCCAT | TTCGACGCACCGGTAGATTA | 5.53 |
| vf0205536817 | ATACACGACGTACGACTGCT | GGGAGGTGTTCGACAGTGTA | 5.53 |
| vf0205547503 | ACAGGATTGAAATACCCTAACACA | TCCCTAAATGTTTGATGCCGT | 5.54 |
| vf0205558302 | AGACCATTAGAGCCCCCATA | GGGGCTAAACCAAAATGACCA | 5.55 |
| vf0205705182 | GTGCAGGTTACCCCCGATTA | TTCTCTATGGACTAGTTGCACAA | 5.7 |
| vf0205722820 | TTAGGCTTAGGCTCATTGGC | TGCATTGCTCAGCTCATGTT | 5.72 |
| vf0205735923 | ATAAACCATCTGCGCACACG | TTGGCGGTGTTCATGGAAAG | 5.73 |
| vf0205742366 | CCTGGATGCGGTTCTTCTTG | ATGGGGAAAAACCGAAAGCC | 5.74 |
| vf0205765597 | ATAGTATGAGGCGGATGCGG | GAAAATTCGCTCCCATGGGG | 5.76 |
| vf0205774130 | GCCTTTGAGCCTCCAAGATG | AGCGAGCCAATGAAAATCCG | 5.77 |
| vf0205774202 | GCCTTTGAGCCTCCAAGATG | AGCGAGCCAATGAAAATCCG | 5.77 |
| vf0205797421 | ATATGCAACCACCGTCTTGC | GGAGATCAACCACCGTCTCA | 5.79 |
| vf0205822727 | ACCATGTTACCCTCTGCCTC | TGGCCATCCATTTTTACGATCA | 5.82 |
| vf0205851262 | GAGTCCTCTTTAGCGGTGGT | ATCCCGACATGTTGGTCACT | 5.85 |
| vf0205858260 | GGAGTCAAATGCTGCTTGCT | GAGGTGTCCAACTTGTGCTG | 5.85 |
| vf0205863221 | CCAAGGCACAGTCTAGGACA | CTGATGCTCCTATACCCGCA | 5.86 |
| vf0205902152 | CAAAGCCATTTCAACGCCAC | TCACTGCAACCACTAGACTACA | 5.9 |
| vf0205943497 | GGACCACAGGAAAAATACAGGA | CCTTTTTAGGCCCCCTTTGA | 5.94 |
| vf0205962045 | TGGTGTTTTTCTGCTGGTCG | CCAACTGCCACAATGTGACA | 5.96 |
| vf0206109126 | CACCAATCGAAAGCTAGCAAA | CGGTGATGATGTTGCATGCT | 6.1 |
| vf0206114052 | CGAGACCAATCGATCCTCCA | TAACCTGCATCTGTGGTGGA | 6.11 |
| vf0206119327 | TCAACCTCAACGAGAAGGGA | ATGTGAAGGCCGTTTTCCTC | 6.11 |
| vf0206201082 | GAGGCTATGAGCTGTCGTCA | TGGCTGCATAAAAATATCCCACA | 6.2 |
| vf0206219603 | GTGTCAATTGCTCTGCCGAT | TCCACGAACATGGTAGCACA | 6.21 |
| vf0206255541 | GGAAGAAAGGTTGCCCAGTG | AGCTCATCCCTTCAAGTGCT | 6.25 |
| vf0206256377 | TTGCAGAAGTCCAGGGATGT | TGCGGGCATGTCTGTACTTA | 6.25 |
| vf0206257411 | ACACATCAATTTCCAGTGACCT | AAGCGTGCCTTGCAGATATG | 6.25 |
| vf0206280178 | TGGTATACCTGACCACATGCT | GGTGACAGACCTCCACCTTT | 6.28 |
Supplemental Table 2 InDel markers on chromosome 2 covering flanking region from 5.50 to 6.28 Mb.
| Marker | Forward primers (5´-3´) | Reverse primers (5´-3´) | Position (Mb) |
|---|---|---|---|
| vf0205500813 | CGGGTCGACTTGCCTGAATA | AGAAATGGCGTTGGAAGCAG | 5.5 |
| vf0205520833 | GTCACGCCGATATCGTCAAC | ACGTCATCATCCACTAACAAGT | 5.52 |
| vf0205535398 | ACCATAATGATTCCGGCCAT | TTCGACGCACCGGTAGATTA | 5.53 |
| vf0205536817 | ATACACGACGTACGACTGCT | GGGAGGTGTTCGACAGTGTA | 5.53 |
| vf0205547503 | ACAGGATTGAAATACCCTAACACA | TCCCTAAATGTTTGATGCCGT | 5.54 |
| vf0205558302 | AGACCATTAGAGCCCCCATA | GGGGCTAAACCAAAATGACCA | 5.55 |
| vf0205705182 | GTGCAGGTTACCCCCGATTA | TTCTCTATGGACTAGTTGCACAA | 5.7 |
| vf0205722820 | TTAGGCTTAGGCTCATTGGC | TGCATTGCTCAGCTCATGTT | 5.72 |
| vf0205735923 | ATAAACCATCTGCGCACACG | TTGGCGGTGTTCATGGAAAG | 5.73 |
| vf0205742366 | CCTGGATGCGGTTCTTCTTG | ATGGGGAAAAACCGAAAGCC | 5.74 |
| vf0205765597 | ATAGTATGAGGCGGATGCGG | GAAAATTCGCTCCCATGGGG | 5.76 |
| vf0205774130 | GCCTTTGAGCCTCCAAGATG | AGCGAGCCAATGAAAATCCG | 5.77 |
| vf0205774202 | GCCTTTGAGCCTCCAAGATG | AGCGAGCCAATGAAAATCCG | 5.77 |
| vf0205797421 | ATATGCAACCACCGTCTTGC | GGAGATCAACCACCGTCTCA | 5.79 |
| vf0205822727 | ACCATGTTACCCTCTGCCTC | TGGCCATCCATTTTTACGATCA | 5.82 |
| vf0205851262 | GAGTCCTCTTTAGCGGTGGT | ATCCCGACATGTTGGTCACT | 5.85 |
| vf0205858260 | GGAGTCAAATGCTGCTTGCT | GAGGTGTCCAACTTGTGCTG | 5.85 |
| vf0205863221 | CCAAGGCACAGTCTAGGACA | CTGATGCTCCTATACCCGCA | 5.86 |
| vf0205902152 | CAAAGCCATTTCAACGCCAC | TCACTGCAACCACTAGACTACA | 5.9 |
| vf0205943497 | GGACCACAGGAAAAATACAGGA | CCTTTTTAGGCCCCCTTTGA | 5.94 |
| vf0205962045 | TGGTGTTTTTCTGCTGGTCG | CCAACTGCCACAATGTGACA | 5.96 |
| vf0206109126 | CACCAATCGAAAGCTAGCAAA | CGGTGATGATGTTGCATGCT | 6.1 |
| vf0206114052 | CGAGACCAATCGATCCTCCA | TAACCTGCATCTGTGGTGGA | 6.11 |
| vf0206119327 | TCAACCTCAACGAGAAGGGA | ATGTGAAGGCCGTTTTCCTC | 6.11 |
| vf0206201082 | GAGGCTATGAGCTGTCGTCA | TGGCTGCATAAAAATATCCCACA | 6.2 |
| vf0206219603 | GTGTCAATTGCTCTGCCGAT | TCCACGAACATGGTAGCACA | 6.21 |
| vf0206255541 | GGAAGAAAGGTTGCCCAGTG | AGCTCATCCCTTCAAGTGCT | 6.25 |
| vf0206256377 | TTGCAGAAGTCCAGGGATGT | TGCGGGCATGTCTGTACTTA | 6.25 |
| vf0206257411 | ACACATCAATTTCCAGTGACCT | AAGCGTGCCTTGCAGATATG | 6.25 |
| vf0206280178 | TGGTATACCTGACCACATGCT | GGTGACAGACCTCCACCTTT | 6.28 |
| [1] | Bai B, Wu J, Sheng W T, Zhou B, Zhou L J, Zhuang W, Yao D P, Deng Q Y.2015. Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress.Int J Mol Sci, 16(5): 11398-11416. |
| [2] | Borkakati R R, Virmani S S.1996. Genetics of thermosensitive genic male sterility in rice.Euphytica, 88(1): 1-7. |
| [3] | Bruinsma J.2003. World Agriculture: Towards 2015/2030. London, UK: Earthscan. |
| [4] | Cao L Y, Zhan X D.2014. Chinese experiences in breeding three-line, two-line and super hybrid rice. In: Yan W G, Bao J S. Rice: Germplasm, Genetics and Improvement. Intech: 279-308. |
| [5] | Chueasiri C, Chunthong K, Pitnjam K, Chakhonkaen S, Sangarwut N, Sangsawang K, Suksangpanomrung M, Michaelson L V, Napier J A, Muangprom A.2014. Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development.PLoS One, 9(9): e106386. |
| [6] | Chunthong K, Pitnjam K, Chakhonkaen S, Sangarwut N, Panyawut N, Wasinanon T, Ukoskit K, Muangprom A.2017. Differential drought responses in F-box gene expression and grain yield between two rice groups with contrasting drought tolerance.J Plant Growth Regul, 36: 970-982. |
| [7] | Ding J H, Lu Q, Ouyang Y D, Mao H L, Zhang P B, Yao J L, Xu C G, Li X H, Xiao J H, Zhang Q F.2012a. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice.Proc Natl Acad Sci USA, 109(7): 2654-2659. |
| [8] | Ding J H, Shen J Q, Mao H L, Xie W B, Li X H, Zhang Q F.2012b. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice.Mol Plant, 5(6): 1210-1216. |
| [9] | Dong N V, Subudhi P K, Luong P N, Quang V D, Quy T D, Zheng H G, Wang B, Nguyen H T.2000. Molecular mapping of a rice gene conditioning thermo-sensitive genic male sterility using AFLP, RFLP and SSR techniques.Theor Appl Genet, 100(5): 727-734. |
| [10] | FAO.2018. FAO Statistical Pocketbook 2018: World Food and Agriculture. Rome: FAO. |
| [11] | Hussain A J, Ali J, Siddiq E A, Gupta V S, Reddy U K, Ranjekar P K.2011. Mapping of tms8 gene for temperature-sensitive genic male sterility (TGMS) in rice(Oryza sativa L.). Plant Breeding, 131(1): 42-47. |
| [12] | Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi A K, Khurana J P.2007. F-Box proteins in rice: Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress.Plant Physiol, 143(4): 1467-1483. |
| [13] | Jia J H, Zhang D S, Li C Y, Qu X P, Wang S W, Chamarerk V, Nguyen H T, Wang B Y.2001. Molecular mapping of the reverse thermosensitive genic male-sterile gene (rtms1) in rice. Theor Appl Genet, 103(4): 607-612. |
| [14] | Jiang D G, Lu S, Zhou H, Wu X J, Zhuang C X, Liu Y G, Mei M T.2006. Mapping of the rice (Oryza sativa L.) thermo-sensitive genic male sterile gene tms5 with EST and SSR markers. Chin Sci Bull, 51(4): 417-420. |
| [15] | Jung K H, Han M J, Lee Y S, Kim Y W, Hwang I, Kim M J, Kim Y K, Nahm B H, An G.2005. Rice undeveloped tapetum1 is a major regulator of early tapetum development. Plant Cell, 17(10): 2705-2722. |
| [16] | Jung K H, Han M J, Lee D Y, Lee Y S, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim Y W, Hwang I, An G.2006. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell, 18(11): 3015-3032. |
| [17] | Lang N T, Subudhi P K, Virmani S S, Brar D S, Khush G S, Li Z, Huang N.1999. Development of PCR-based markers for thermosensitive genetic male sterility gene tms3(t) in rice (Oryza satvai L.). Hereditas, 131(2): 121-127. |
| [18] | Lee D S, Chen L J, Suh H S.2005. Genetic characterization and fine mapping of a novel thermo-sensitive genic male-sterile genetms6 in rice(Oryza sativa L.). Theor Appl Genet, 111(7): 1271-1277. |
| [19] | Li R B, Pandey M P, Sharma P, Ballabh G.2005. Inheritance of thermosensitive genic male sterility in rice (Oryza sativa L.). Curr Sci, 88(11): 1809-1815. |
| [20] | Li N, Zhang D S, Liu H S, Yin C S, Li X X, Liang W Q, Yuan Z, Xu B, Chu H W, Wang J, Wen T Q, Huang H, Luo D, Ma H, Zhang D B.2006. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development.Plant Cell, 18(11): 2999-3014. |
| [21] | Liu N, Shan Y, Wang F P, Xu C G, Peng K M, Li X H, Zhang Q F.2001. Identification of an 85-kb DNA fragment containing pms1, a locus for photoperiod-sensitive genic male sterility in rice. Mol Genet Genom, 266(2): 271-275. |
| [22] | Livak K J, Schmittgen T D.2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method.Methods, 25(4): 402-408. |
| [23] | Lopez M T, Toojinda T, Vanavichit A, Tragoonrung S.2003. Microsatellite markers flanking the tms2 gene facilitated tropical TGMS rice line development. Crop Sci, 43(6): 2267-2271. |
| [24] | Matthayatthaworn W, Sripichitt P, Phumichai C, Rungmekarat S, Uckarach S, Sreewongchai T.2011. Development of specific simple sequence repeat (SSR) markers for non-pollen type thermo-sensitive genic male sterile gene in rice (Oryza sativa L.). Afr J Biotechnol, 10: 16437-16442. |
| [25] | McCouch S R, Teytelman L, Xu Y, Lobos K B, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L.2002. Development and mapping of 2240 new SSR markers for rice ( Oryza sativa L.). DNA Res, 9(6): 199-207. |
| [26] | Murray M G, Thompson W F.1980. Rapid isolation of high molecular weight plant DNA.Nucl Acid Res, 8(19): 4321-4325. |
| [27] | Nas T M S, Sanchez D L, Diaz G Q, Mendioro M S, Vermani S S.2005. Pyramiding of thermosensitive genetic male sterility (TGMS) genes and identification of a candidate tms5 gene in rice. Euphytica, 145: 67-75. |
| [28] | Nonomura K I, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N.2003. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell, 15(8): 1728-1739. |
| [29] | Peng H F, Zhang Z F, Wu B, Chen X H, Zhang G Q, Zhang Z M, Wan B H, Lu Y P.2008. Molecular mapping of two reverse photoperiod-sensitive genic male sterility genes (rpms1 and rpms2) in rice(Oryza sativa L.). Theor Appl Genet, 118(1): 77-83. |
| [30] | Peng H F, Chen X H, Lu Y P, Peng Y F, Wan B H, Chen N D, Wu B, Xin S P, Zhang G Q.2010. Fine mapping of a gene for non-pollen type thermosensitive genic male sterility in rice (Oryza sativa L.). Theor Appl Genet, 120(5): 1013-1020. |
| [31] | Pitnjam K, Chakhonkaen S, Toojinda T, Muangprom A.2008. Identification of a deletion in tms2 and development of gene-based markers for selection. Planta, 228(5): 813-822. |
| [32] | Qi Y B, Liu Q L, Zhang L, Mao B Z, Yan D W, Jin Q S, He Z H.2014. Fine mapping and candidate gene analysis of the novel thermo sensitive genic male sterility tms9-1 gene in rice. Theor Appl Genet, 127(5): 1173-1182. |
| [33] | Sharma M, Pandey G K.2016. Expansion and function of repeat domain proteins during stress and development in plants.Front Plant Sci, 6: 1218. |
| [34] | Sheng Z H, Wei X J, Shao G N, Chen M L, Song J, Tang S Q, Luo J, Hu Y C, Hu P S, Chen L Y.2013. Genetic analysis and fine mapping of tms9, a novel thermosensitive genic male-sterile gene in rice(Oryza sativa L.). Plant Breeding, 132(2): 159-164. |
| [35] | Shin S B, Golovkin M, Reddy A S N.2014. A pollen-specific calmodulin-binding protein, NPG1, interacts with putative pectate lyases.Sci Rep, 4: 5263. |
| [36] | Small I D, Peeters N.2000. The PPR motif: A TPR-related motif prevalent in plant organellar proteins.Trends Biochem Sci, 25(2): 45-47. |
| [37] | Subudhi P K, Borkakati R P, Virmani S S, Huang N.1997. Molecular mapping of a thermo-sensitive genetic male-sterility gene in rice using bulked segregant analysis.Genome, 40(2): 188-194. |
| [38] | Thai Rice Exporters Association. 2018. World Rice Production and Ending Stocks.. |
| [39] | USDA. 2016. World Agricultural Production. . |
| [40] | Virmani S S.2003. Advances in hybrid rice research and development in the tropics. In: Virmani S S, Mao C X, Hardy B. Hybrid Rice for Food Security, Poverty Alleviation, and Environmental Protection . Proceedings of the 4th International Symposium on Hybrid Rice, Hanoi, Vietnam. Los Baños,the Philippines: International Rice Research Institute: 7-20. |
| [41] | Wang B Y, Xu W W, Wang J Z, Wu W, Zheng H G, Yang Z Y, Ray J D, Nguyen H T.1995. Tagging and mapping the thermo-sensitive genic male-sterile gene in rice (Oryza sativa) with molecular markers. Theor Appl Genet, 91: 1111-1114. |
| [42] | Wang Y G, Xing Q H, Deng Q Y, Liang F S, Yuan L P, Weng M L, Wang B.2003. Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor Appl Genet, 107(5): 917-921. |
| [43] | Xu J J, Wang B H, Wu Y H, Du P N, Wang J, Wang M, Yi C D, Gu M H, Liang G H.2011. Fine mapping and candidate gene analysis of ptgms2-1, the photoperiod-thermo-sensitive genic male sterile gene in rice(Oryza sativa L.). Theor Appl Genet, 122(2): 365-372. |
| [44] | Yamaguchi Y, Ikeda R, Hirasawa H, Minami M, Ujihara A.1997. Linkage analysis of thermo-sensitive genic male sterility gene tms-2 in rice(Oryza sativa L.). Breeding Sci, 47: 371-373. |
| [45] | Yang Q K, Liang C Y, Zhuang W, Li J, Deng H B, Deng Q Y, Wang B.2007. Characterization and identification of the candidate gene of rice thermo-sensitive genic male sterile gene tms5 by mapping. Planta, 225(2): 321-330. |
| [46] | Yang Y J, Ma C, Xu Y J, Wei Q, Imtiaz M, Lan H B, Gao S, Cheng L N, Wang M Y, Fei Z J, Hong B, Gao J P.2014. A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum by modulating gibberellin biosynthesis.Plant Cell, 26(5): 2038-2054. |
| [47] | Yang J, Chen X R, Zhu C L, Peng X S, He X P, Fu J R, Ouyang L J, Bian J M, Hu L F, Sun X T, Xu J, He H H.2015. RNA-Seq reveals differentially expressed genes of rice (Oryza sativa) spikelet in response to interacting with nitrogen at meiosis stage. BMC Genom, 16: 959. |
| [48] | Yuan L P.1998. Hybrid rice breeding in China. In: Virmani S S, Siddiq E A, Muralidharan K. Advances in Hybrid Rice Technology. Proceedings of the 3rd International Symposium on Hybrid Rice. Hyderabad, India. Manila, the Philippines: International Rice Research Institute: 27-33. |
| [49] | Zhang Q F, Shen B Z, Dai X K, Mei M H, Saghai Maroof M A, Li Z B.1994. Using bulked extremes and recessive classes to map genes for photoperiod-sensitive genic male sterility in rice.Proc Natl Acad Sci USA, 91(18): 8675-8679. |
| [50] | Zhang Q S, Li Z, Yang J, Li S Q, Yang D C, Zhu Y G.2012. A calmodulin-binding protein from rice is essential to pollen development.J Plant Biol, 55(1): 8-14. |
| [51] | Zhou Y F, Zhang X Y, Xue Q Z.2011. Fine mapping and candidate gene prediction of the photoperiod and thermo- sensitive genic male sterile gene pms1(t) in rice. J Zhejiang Univ: Sci B, 12(6): 436-447. |
| [52] | Zhou H, Liu Q J, Li J, Jiang D G, Zhou L Y, Wu P, Lu S, Li F, Zhu L Y, Liu Z L, Chen L T, Liu Y G, Zhuang C X.2012. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA.Cell Res, 22(4): 649-660. |
| [53] | Zhou H, Zhou M, Yang Y Z, Li J, Zhu L Y, Jiang D G, Dong J F, Liu Q J, Gu L F, Zhou L Y, Feng M J, Qin P, Hu X C, Song C L, Shi J F, Song X W, Ni E D, Wu X J, Deng Q Y, Liu Z L, Chen M S, Liu Y G, Cao X F, Zhuang C X.2014. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice.Nat Commun, 5: 4884. |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [2] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [3] | Yanchang Luo, Tingchen Ma, Teo Joanne, Zhixiang Luo, Zefu Li, Jianbo Yang, Zhongchao Yin. Marker-Assisted Breeding of Thermo-Sensitive Genic Male Sterile Line 1892S for Disease Resistance and Submergence Tolerance [J]. Rice Science, 2021, 28(1): 89-98. |
| [4] | Jan Mehmood, Shah Gulmeena, Yuqing Huang, Xuejiao Liu, Peng Zheng, Hao Du, Hao Chen, Jumin Tu. Development of Heat Tolerant Two-Line Hybrid Rice Restorer Line Carrying Dominant Locus of OsHTAS [J]. Rice Science, 2021, 28(1): 99-108. |
| [5] | Pathaichindachote Wanwarang, Panyawut Natjaree, Sikaewtung Kannika, Patarapuwadol Sujin, Muangprom Amorntip. Genetic Diversity and Allelic Frequency of Selected Thai and Exotic Rice Germplasm Using SSR Markers [J]. Rice Science, 2019, 26(6): 393-403. |
| [6] | Ngangkham Umakanta, Kumar Parida Swarup, Kumar Singh Ashok, Mohapatra Trilochan. Differential RNA Editing of Mitochondrial Genes in WA-Cytoplasmic Based Male Sterile Line Pusa 6A, and Its Maintainer and Restorer Lines [J]. Rice Science, 2019, 26(5): 282-289. |
| [7] | Donde Ravindra, Kumar Jitendra, Gouda Gayatri, Kumar Gupta Manoj, Mukherjee Mitadru, Yasin Baksh Sk, Mahadani Pradosh, Kumar Sahoo Khirod, Behera Lambodar, Kumar Dash Sushanta. Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers [J]. Rice Science, 2019, 26(4): 239-247. |
| [8] | F. Aala Jr Wilson, B. Gregorio Glenn. Morphological and Molecular Characterization of Novel Salt-Tolerant Rice Germplasms from the Philippines and Bangladesh [J]. Rice Science, 2019, 26(3): 178-188. |
| [9] | Wenqiang Liu, Xiaowu Pan, Yongchao Li, Yonghong Duan, Jun Min, Sanxiong Liu, Licheng Liu, Xinnian Sheng, Xiaoxiang Li. Identification of QTLs and Validation of qCd-2 Associated with Grain Cadmium Concentrations in Rice [J]. Rice Science, 2019, 26(1): 42-49. |
| [10] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| [11] | Haritha G., P. M. Swamy B., L. Naik M., Jyothi B., Divya B., Malathi S., Sarla N.. Yield Traits and Associated Marker Segregation in Elite Introgression Lines Derived from O. sativa × O. nivara [J]. Rice Science, 2018, 25(1): 19-31. |
| [12] | Thi Nguyen Hue, Hong Vu Quang, Van Mai Tan, Thi Nguyen Thu, Duc Vu Lam, Thanh Nguyen Tung, Viet Nguyen Long, Thu Thi Vu Hien, Thi Nong Hue, Nguyen Dinh Trung, Toshitsugu Nakano, Van Vu Liet. Marker-Assisted Selection of Xa21 Conferring Resistance to Bacterial Leaf Blight in indica Rice Cultivar LT2 [J]. Rice Science, 2018, 25(1): 52-56. |
| [13] | Naga Bheema Lingeswara Reddy Inja, Kim Sung-Mi, Kim Beom-Ki, Yoon In-Sun, Kwon Taek-Ryoun. Identification of Rice Accessions Associated with K+/Na+ Ratio and Salt Tolerance Based on Physiological and Molecular Responses [J]. Rice Science, 2017, 24(6): 360-364. |
| [14] | Yan Liang, Bai-yuan Yan, Yun-liang Peng, Zhi-juan Ji, Yu-xiang Zeng, Han-lin Wu, Chang-deng Yang. Molecular Screening of Blast Resistance Genes in Rice Germplasms Resistant to Magnaporthe oryzae [J]. Rice Science, 2017, 24(1): 41-47. |
| [15] | D. Chowdhury A., Haritha G., Sunitha T., L. Krishnamurthy S., Divya B., Padmavathi G., Ram T., Sarla N.. Haplotyping of Rice Genotypes Using Simple Sequence Repeat Markers Associated with Salt Tolerance [J]. Rice Science, 2016, 23(6): 317-325. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||