
Rice Science ›› 2019, Vol. 26 ›› Issue (5): 282-289.DOI: 10.1016/j.rsci.2019.08.002
• Research Papers • Previous Articles Next Articles
Ngangkham Umakanta1,2( ), Kumar Parida Swarup1,3, Kumar Singh Ashok4, Mohapatra Trilochan1,5(
), Kumar Parida Swarup1,3, Kumar Singh Ashok4, Mohapatra Trilochan1,5( )
)
Received:2018-07-19
Accepted:2018-10-10
Online:2019-09-28
Published:2019-05-24
Ngangkham Umakanta, Kumar Parida Swarup, Kumar Singh Ashok, Mohapatra Trilochan. Differential RNA Editing of Mitochondrial Genes in WA-Cytoplasmic Based Male Sterile Line Pusa 6A, and Its Maintainer and Restorer Lines[J]. Rice Science, 2019, 26(5): 282-289.
Add to citation manager EndNote|Ris|BibTeX
| Gene | Biological function | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size (bp) |
|---|---|---|---|---|
| 18SrRNA | Ribosomal subunit | GTGTTGCTGAGACATGCGCC | ATATGGCGCAAGACGATTCC | 749 |
| atp6 | Subunit of ATPase | ATGGGTTTGAATCAGAGAGA | ATTCAATTATGAAATTACTC | 999 |
| atp9 | Subunit of ATPase | GGTGTGGTGTTCAGTCTACC | GGGCCTCGTATCTCTATTTG | 406 |
| cobII | Apocytochrome B | AAGGAACCAACGATTCTCTC | CGGTCGAAAACTTGAACTAC | 1 001 |
| coxI | Cytochrome C oxidase subunit | TATTACCAGCCATTCTGGAG | CTACGAAGAAACGACGAATC | 1 008 |
| coxIII | Cytochrome C oxidase subunit | GGAGAGGGCATGATAAAGAC | AAATAGTGGAGGGTGCTTG | 810 |
| nadI | Subunit of NADH dehydrogenase | ATACACCAGGGCAACTAATG | AGGGAGTAGGGTGAGTAAGC | 790 |
| rps3 | Ribosomal subunit | AAACCCTCTCTAAGGTGGAG | AGATAGTCACCCATCACACG | 1 002 |
Table 1 Eight rice mitochondrial genes and their biological function used in this study.
| Gene | Biological function | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size (bp) |
|---|---|---|---|---|
| 18SrRNA | Ribosomal subunit | GTGTTGCTGAGACATGCGCC | ATATGGCGCAAGACGATTCC | 749 |
| atp6 | Subunit of ATPase | ATGGGTTTGAATCAGAGAGA | ATTCAATTATGAAATTACTC | 999 |
| atp9 | Subunit of ATPase | GGTGTGGTGTTCAGTCTACC | GGGCCTCGTATCTCTATTTG | 406 |
| cobII | Apocytochrome B | AAGGAACCAACGATTCTCTC | CGGTCGAAAACTTGAACTAC | 1 001 |
| coxI | Cytochrome C oxidase subunit | TATTACCAGCCATTCTGGAG | CTACGAAGAAACGACGAATC | 1 008 |
| coxIII | Cytochrome C oxidase subunit | GGAGAGGGCATGATAAAGAC | AAATAGTGGAGGGTGCTTG | 810 |
| nadI | Subunit of NADH dehydrogenase | ATACACCAGGGCAACTAATG | AGGGAGTAGGGTGAGTAAGC | 790 |
| rps3 | Ribosomal subunit | AAACCCTCTCTAAGGTGGAG | AGATAGTCACCCATCACACG | 1 002 |
| Gene | Pusa 6A | Pusa 6B | PRR78 | TPPE | TF (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS | NI | ND | NT | F (%) | NS | NI | ND | NT | F (%) | NS | NI | ND | NT | F (%) | |||||
| 18SrRNA | 2 | - | - | 2 | 0.27 | 2 | - | 1 | 3 | 0.4 | 29 | 7 | - | 36 | 4.8 | 41 | 5.47 | ||
| atp6 | 6 | 2 | - | 8 | 0.8 | 31 | 3 | 2 | 36 | 3.6 | 11 | 1 | - | 12 | 1.2 | 52 | 5.21 | ||
| atp9 | - | 1 | - | 1 | 0.24 | 2 | 1 | 5 | 8 | 1.97 | - | - | 1 | 1 | 0.24 | 8 | 1.97 | ||
| cobII | 7 | 2 | - | 9 | 0.9 | 20 | 3 | 1 | 24 | 2.4 | 12 | 2 | - | 14 | 1.4 | 36 | 3.6 | ||
| coxI | - | - | - | - | 0 | 6 | - | - | 6 | 0.6 | - | - | - | - | 0 | 6 | 0.6 | ||
| coxIII | 7 | 3 | 1 | 11 | 1.36 | 10 | 3 | 2 | 15 | 1.85 | 9 | 1 | 1 | 11 | 1.36 | 23 | 2.84 | ||
| nadI | - | 1 | - | 1 | 0.12 | 2 | - | - | 2 | 0.25 | 19 | 2 | 11 | 32 | 4.05 | 35 | 4.43 | ||
| rps3 | 6 | 4 | - | 10 | 1 | 13 | - | 6 | 19 | 1.89 | 2 | 1 | - | 3 | 0.3 | 29 | 3.19 | ||
| Total | 28 | 13 | 1 | 42 | 0.62 | 86 | 10 | 17 | 113 | 1.67 | 82 | 14 | 13 | 109 | 1.61 | 230 | 3.39 | ||
Table 2 Statistics of eight mitochondrial genes in Pusa 6A, Pusa 6B and PRR78.
| Gene | Pusa 6A | Pusa 6B | PRR78 | TPPE | TF (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS | NI | ND | NT | F (%) | NS | NI | ND | NT | F (%) | NS | NI | ND | NT | F (%) | |||||
| 18SrRNA | 2 | - | - | 2 | 0.27 | 2 | - | 1 | 3 | 0.4 | 29 | 7 | - | 36 | 4.8 | 41 | 5.47 | ||
| atp6 | 6 | 2 | - | 8 | 0.8 | 31 | 3 | 2 | 36 | 3.6 | 11 | 1 | - | 12 | 1.2 | 52 | 5.21 | ||
| atp9 | - | 1 | - | 1 | 0.24 | 2 | 1 | 5 | 8 | 1.97 | - | - | 1 | 1 | 0.24 | 8 | 1.97 | ||
| cobII | 7 | 2 | - | 9 | 0.9 | 20 | 3 | 1 | 24 | 2.4 | 12 | 2 | - | 14 | 1.4 | 36 | 3.6 | ||
| coxI | - | - | - | - | 0 | 6 | - | - | 6 | 0.6 | - | - | - | - | 0 | 6 | 0.6 | ||
| coxIII | 7 | 3 | 1 | 11 | 1.36 | 10 | 3 | 2 | 15 | 1.85 | 9 | 1 | 1 | 11 | 1.36 | 23 | 2.84 | ||
| nadI | - | 1 | - | 1 | 0.12 | 2 | - | - | 2 | 0.25 | 19 | 2 | 11 | 32 | 4.05 | 35 | 4.43 | ||
| rps3 | 6 | 4 | - | 10 | 1 | 13 | - | 6 | 19 | 1.89 | 2 | 1 | - | 3 | 0.3 | 29 | 3.19 | ||
| Total | 28 | 13 | 1 | 42 | 0.62 | 86 | 10 | 17 | 113 | 1.67 | 82 | 14 | 13 | 109 | 1.61 | 230 | 3.39 | ||
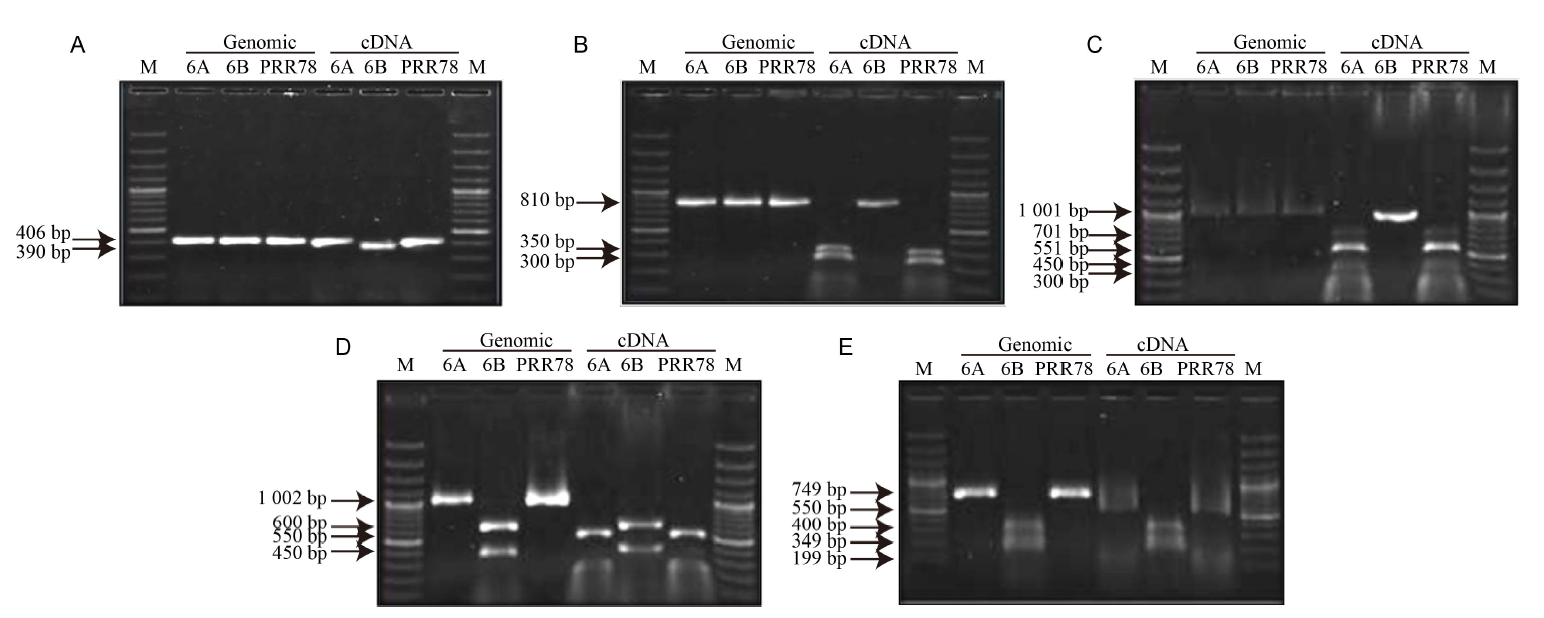
Fig. 1. Validation of RNA editing in mitochondrial genes by cleaved amplified polymorphism sequence analysis. M, 100 bp DNA ladder plus; 6A, CMS line Pusa 6A; 6B, Maintainer line Pusa 6B; PRR78, Restorer line PRR78.The recognition sites of DpnI (GATC) for atp9 (A), HpyCH4IV (ACGT) for coxIII (B), Tsp509I (AATT) for cobII (C), BfaI (CTAG) for rps3 (D) and AluI (AGCT) for 18SrRNA (E) were affected by SNPs T/G, C/A, T/A, G/C and A/T as predicted by SNP2CAPS, respectively. The digested PCR fragments were separated in 1.2% agarose gel.
| [1] | Benne R, van den Burg J, Brakenhoff J P, Sloof P, van Boom J H, Tromp M C.1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell, 46(6): 819-826. |
| [2] | Cattaneo R.1991. Different types of messenger RNA editing.Annu Rev Genet, 25: 71-88. |
| [3] | Chakraborty A, Mitra J, Bhattacharyya J, Pradhan S, Sikdar N, Das S, Chakraborty S, Kumar S, Lakhanpaul S, Sen S K.2015. Transgenic expression of an unedited mitochondrial orfB gene product from wild abortive (WA) cytoplasm of rice(Oryza sativa L.) generates male sterility in fertile rice lines. Planta, 241: 1463-1479. |
| [4] | Chateigner-Boutin A L, Ramos-Vega M, Guevara-Garcia A, Andres C, de la Luz Gutierrez-Nava M, Cantero A, Delannoy E, Jiménez L F, Lurin C, Small I, León P.2008. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J, 56(4): 590-602. |
| [5] | Covello P S, Gray M W.1989. RNA editing in plant mitochondria.Nature, 341: 662-666. |
| [6] | Covello P S, Gray M W.1990. Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria.Nucl Acids Res, 18(17): 5189-5196. |
| [7] | Das S, Sen S, Chakraborty A, Chakraborti P, Maiti M K, Basu A, Basu D, Sen S K.2010. An unedited 1.1 kb mitochondrial orfB gene transcript in the wild abortive cytoplasmic male sterility WA-CMS system of Oryza sativa L. subsp. indica. BMC Plant Biol, 10: 39. |
| [8] | Dewey R E, Levings C S, Timothy D H.1986. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male sterile cytoplasm. Cell, 44(3): 439-449. |
| [9] | Doyle J J, Doyle J L.1990. Isolation of plant DNA from fresh tissue.Focus, 12: 13-15. |
| [10] | Giege P, Brennicke A.1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA, 96: 15324-15329. |
| [11] | Graves P V, Bégu D, Velours J, Neau E, Belloc F, Litvak S, Araya A.1990. Direct protein sequencing of wheat mitochondrial ATP-synthase subunit 9 confirms RNA editing in plants.J Mol Biol, 214(1): 1-6. |
| [12] | Gray M W, Hanic-Joyce P J, Covello P S.1992. Transcription, processing and editing in plant mitochondria.Annu Rev Plant Physiol Plant Mol Biol, 43: 145-175. |
| [13] | Gray M W, Covello P S.1993. RNA editing in plant mitochondria and chloroplasts.FASEB J, 7: 64-71. |
| [14] | Grosskopf D, Mulligan R M.1996. Developmental and tissue specificity of RNA editing in mitochondria of suspension cultured maize cells and seedlings.Curr Genet, 29(6): 556-563. |
| [15] | Handa H, Gualberto J M, Grienenberger J M.1995. Characterization of the mitochondrial orfB gene and its derivative, orf224, a chimeric open reading frame specific to one mitochondrial genome the Polima CMS in rapeseed Brassica napus L. Curr Genet, 28(6): 546-552. |
| [16] | Hanson M R, Sutton C A, Lu B W.1996. Plant organelle gene expression: Altered by RNA editing.Trends Plant Sci, 1(2): 57-64. |
| [17] | Hernould M, Suharsono S, Litvak S, Araya A, Mouras A.1993. Male sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc Natl Acad Sci USA, 90(6): 2370-2374. |
| [18] | Hiesel R, Wissinger B, Schuster W, Brennicke A.1989. RNA editing in plant mitochondria.Science, 246: 1632-1634. |
| [19] | Horn R, Gupta K J, Colombo N.2014. Mitochondrion role in molecular basis of cytoplasmic male sterility.Mitochondrion, 19: 198-205. |
| [20] | Howad W, Kempken F.1997. Cell type-specific loss of atp6 RNA editing in cytoplasmic male sterile Sorghum bicolor. Proc Natl Acad Sci USA, 94: 11090-11095. |
| [21] | Hu J H, Yi R, Zhang H Y, Ding Y.2013. Nucleo-cytoplasmic interactions affect RNA editing of cox2, atp6, and atp9 in alloplasmic male sterile rice(Oryza sativa L.) lines. Mitochondrion, 13(2): 87-95. |
| [22] | Iwabuchi M, Kyozuka J, Shimamoto K.1993. Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male-sterile rice. EMBO J, 12(4): 1437-1446. |
| [23] | Ji J J, Huang W, Li D W, Yin Y X, Chai W G, Gong Z H.2014. A CMS-related gene,Ψatp6-2, causes increased ATP hydrolysis activity of the mitochondrial F1Fo-ATP synthase and induces male sterility in pepper(Capsicum annuum L). Plant Mol Biol Rep, 32(4): 888-899. |
| [24] | Kianian P M A, Kianian S F.2014. Mitochondrial dynamics and the cell cycle.Front Plant Sci, 5: 222. |
| [25] | Liew Y J, Li Y, Baumgarten S, Voolstra C R, Aranda M.2017. Condition-specific RNA editing in the coral symbiontSymbiodinium microadriaticum. PLoS Genet, 13(2): e1006619. |
| [26] | Liu Z L, Xu H, Guo J X, Liu Y G.2007. Structural and expression variations of the mitochondrial genome conferring the WA type of CMS in rice.J Integr Plant Biol, 49(6): 908-914. |
| [27] | Luo D P, Xu H, Liu Z L, Guo J X, Li H Y, Chen L T, Fang C, Zhang Q Y, Bai M, Yao N, Wu H, Wu H, Ji C H, Zheng H Q, Chen Y L, Ye S, Li X Y, Zhao X C, Li R Q, Liu Y G.2013. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice.Nat Genet, 45(5): 573-577. |
| [28] | Mower J P, Palmer J D.2006. Patterns of partial RNA editing in mitochondrial genes ofBeta vulgaris. Mol Genet Genomics, 276(3): 285-293. |
| [29] | Ngangkham U, Parida S K, De S, Kumar A R, Singh A K, Singh N K, Mohapatra T.2010. Genic markers for wild abortive WA cytoplasm based male sterility and its fertility restoration in rice.Mol Breeding, 26: 275-292. |
| [30] | Picardi E, Horner D S, Chiara M, Schiavon R, Valle G, Pesole G.2010. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing.Nucleic Acids Res, 38(14): 4755-4767. |
| [31] | Schuster W, Hiessel R, Wissiinger B, Brennicke A.1990. RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U to C transition. Mol Cell Biol, 10(5): 2428-2431. |
| [32] | Smith H C, Gott J M, Hanson M R.1997. A guide to RNA editing.RNA, 3(10): 1105-1123. |
| [33] | Stuart K, Allen T E, Heideman S, Seiwert S D.1997. RNA editing of kinetoplast protozoa.Microl Mol Biol Rev, 61(1): 105-120. |
| [34] | Thiel T, Kota R, Grosse I, Stein N, Graner A.2004. SNP2CAPS: A SNP and INDEL analysis tool for CAPS marker development.Nucleic Acids Res, 32(1): 5. |
| [35] | Wu B, Chen H M, Shao J J, Zhang H, Wu K, Liu C.2017. Identification of symmetrical RNA editing events in the mitochondria of Salvia miltiorrhiza by strand specific RNA sequencing. Sci Rep, 7: 42250. |
| [36] | Young E G, Hanson M R.1987. A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated.Cell, 50(1): 41-49. |
| [37] | Yu J H, Zhao Y X, Qin Y T, Yue B, Zheng Y L, Xiao H L.2013. Discovery of microRNAs associated with the S type cytoplasmic male sterility in maize.J Integr Agric, 12(2): 229-238. |
| [38] | Zehrmann A, van der Merwe J A, Veritskiy D, Brennicke A, Takenaka M.2008. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis. Mitochondrion, 8(4): 319-327. |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [2] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [3] | Shalini Pulipati, Suji Somasundaram, Nitika Rana, Kavitha Kumaresan, Mohamed Shafi, Peter Civáň, Gothandapani Sellamuthu, Deepa Jaganathan, Prasanna Venkatesan Ramaravi, S. Punitha, Kalaimani Raju, Shrikant S. Mantri, R. Sowdhamini, Ajay Parida, Gayatri Venkataraman. Diversity of Sodium Transporter HKT1;5 in Genus Oryza [J]. Rice Science, 2022, 29(1): 31-46. |
| [4] | Yang Lv, Yueying Wang, Jahan Noushin, Haitao Hu, Ping Chen, Lianguang Shang, Haiyan Lin, Guojun Dong, Jiang Hu, Zhenyu Gao, Qian Qian, Yu Zhang, Longbiao Guo. Genome-Wide Association Analysis and Allelic Mining of Grain Shape-Related Traits in Rice [J]. Rice Science, 2019, 26(6): 384-392. |
| [5] | Khlaimongkhon Sudthana, Chakhonkaen Sriprapai, Pitngam Keasinee, Ditthab Khanittha, Sangarwut Numphet, Panyawut Natjaree, Wasinanon Thiwawan, Mongkolsiriwatana Chareerat, Chunwongse Julapark, Muangprom Amorntip. Molecular Markers and Candidate Genes for Thermo-Sensitive Genic Male Sterile in Rice [J]. Rice Science, 2019, 26(3): 147-156. |
| [6] | Yaobin Qin, Peng Cheng, Yichen Cheng, Yue Feng, Derun Huang, Tingxu Huang, Xianjun Song, Jiezheng Ying. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population [J]. Rice Science, 2018, 25(3): 121-131. |
| [7] | Chandra Roy Subhas, Bhasker Reddy Lachagari Vijaya. Assessment of SNP and InDel Variations Among Rice Lines of Tulaipanji x Ranjit [J]. Rice Science, 2017, 24(6): 336-348. |
| [8] | S. M. Masuduzzaman A., Maksudul Haque Md., K. M. Shamsuddin A., A. Salam M., Ansar Ali Md.. Haplotype Diversity at Sub1 Locus and Allelic Distribution Among Rice Varieties of Tide and Flood Prone Areas of South-East Asia [J]. Rice Science, 2017, 24(4): 198-206. |
| [9] | Hong-guang Xie, Jia-huang Jiang, Yan-mei Zheng, Yong-sheng Zhu, Fang-xi Wu, Xi Luo, Qiu-hua Cai, Jian-fu Zhang, Hua-an Xie. Development of Hybrid Rice Variety FY7206 with Blast Resistance Gene Pid3 and Cold Tolerance Gene Ctb1 [J]. Rice Science, 2016, 23(5): 266-273. |
| [10] | El-Namaky Raafat, Sedeek Saber, Dea Moukoumbi Yonnelle, Ortiz Rodomiro, Manneh Baboucarr. Microsatellite-Aided Screening for Fertility Restoration Genes (Rf) Facilitates Hybrid Improvement [J]. Rice Science, 2016, 23(3): 160-164. |
| [11] | Guan-fu Fu, Cai-xia Zhang, Yong-jie Yang, Jie Xiong, Xue-qin Yang, Xiu-fu Zhang, Qian-yu Jin, Long-xing Tao. Male Parent Plays More Important Role in Heat Tolerance in Three-Line Hybrid Rice [J]. Rice Science, 2015, 22(3): 116-122. |
| [12] | Zhi-yuan Huang, Bing-ran Zhao, Qi-ming Lv, Xi-qin Fu, Ye-yun Xin, Long-ping Yuan. Heterosis Expression of Hybrid Rice in Natural- and Short-Day Length Conditions [J]. Rice Science, 2015, 22(2): 81-88. |
| [13] | J. Arasakesary S., Manonmani S., Pushpam R., Robin S.. New Temperature Sensitive Genic Male Sterile Lines with Better Outcrossing Ability for Production of Two-Line Hybrid Rice [J]. Rice Science, 2015, 22(1): 49-52. |
| [14] | LIANG Yan, ZHANG Xue-mei, LI De-qiang, HUANG Fu, HU Pei-song, PENG Yun-liang. Integrated Approach to Control False Smut in Hybrid Rice in Sichuan Province, China [J]. RICE SCIENCE, 2014, 21(6): 354-360. |
| [15] | Mohammad H. FOTOKIAN, Kayvan AGAHI. Biplot Analysis of Genotype by Environment for Cooking Quality in Hybrid Rice: A Tool for Line × Tester Data [J]. RICE SCIENCE, 2014, 21(5): 282-287. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||