
Rice Science ›› 2021, Vol. 28 ›› Issue (3): 243-256.DOI: 10.1016/j.rsci.2021.04.004
• Research Paper • Previous Articles Next Articles
Xiaoqin Zeng#, Hui Zhuang#, Qinglan Cheng#, Jun Tang, Fayu Yang, Mingjiang Huang, Ziyi Wang, Zhongcheng Li, Honghui Zhu, Rui Chen, Guanghua He( ), Yunfeng Li(
), Yunfeng Li( )
)
Received:2021-01-12
Accepted:2021-03-01
Online:2021-05-28
Published:2021-05-28
About author:#These authors contributed equally to this work
Xiaoqin Zeng, Hui Zhuang, Qinglan Cheng, Jun Tang, Fayu Yang, Mingjiang Huang, Ziyi Wang, Zhongcheng Li, Honghui Zhu, Rui Chen, Guanghua He, Yunfeng Li. SB1 Encoding RING-Like Zinc-Finger Protein Regulates Branch Development as a Transcription Repressor[J]. Rice Science, 2021, 28(3): 243-256.
Add to citation manager EndNote|Ris|BibTeX
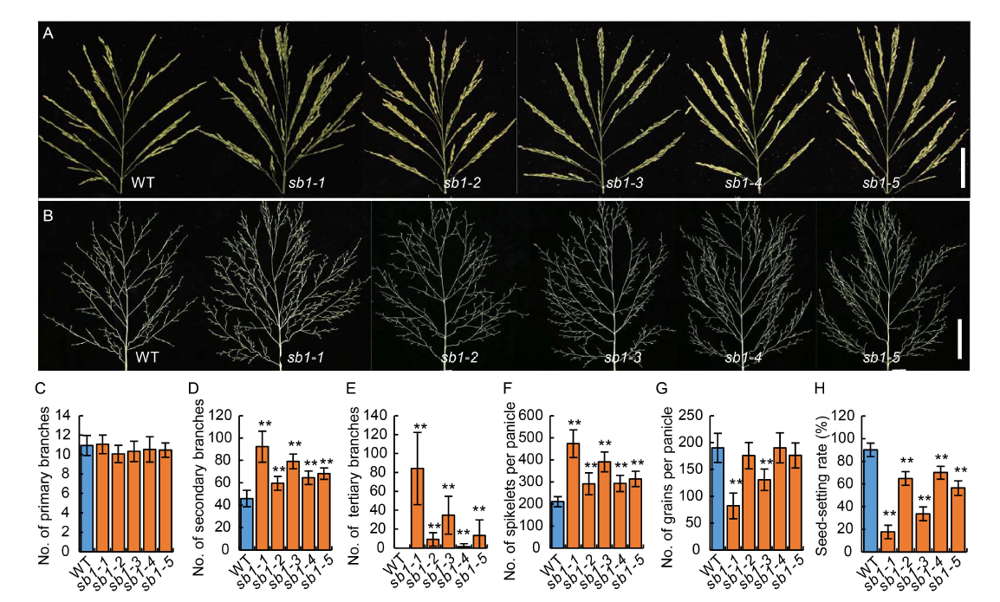
Fig. 1. Phenotypes of panicles in wild type (WT) and sb1-1/2/3/4/5 mutants. A, Panicles of the WT and sb1-1/2/3/4/5 mutants with spikelets on the branches. Bars are 5 cm. B, Panicles of the WT and sb1-1/2/3/4/5 mutants without spikelets on the branches. Bars are 5 cm. C?H, Numbers of primary branches per panicle (C), numbers of secondary branches per panicle (D), numbers of tertiary branches per panicle (E), numbers of spikelets per panicle (F), numbers of grains per panicle (G) and seed-setting rates (H) of WT and sb1-1/2/3/4/5.The wild type plants and sb1-1/2/3/4/5 mutants were grown in standard paddy field under conventional cultivation conditions. Data are Mean ± SD (n = 10). The Student’s t-test was used to generate the P values. **, P < 0.01.
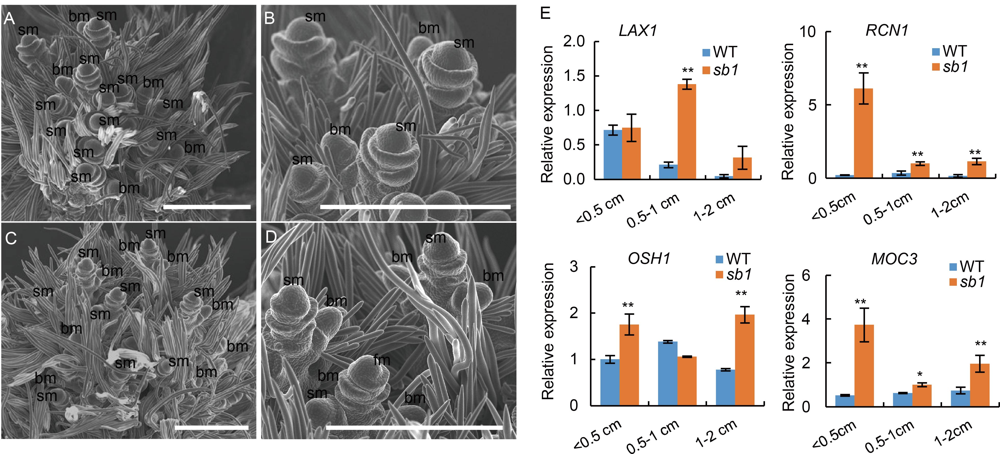
Fig. S1. scanning electron microscopy analysis of young panicles of sb1-1 mutant and wild type. A, Wild type panicles. B, Magnified figure of A. C, sb1-1 panicles. D, Magnified figure of C.E, Expression of LAX1, RCN1, OSH1 and MOC3. bm, branch meristem; sm, spikelet meristem. Bars are 500μm. Data are Mean ± SD (n = 3). The Student’s t-test was used to generate the P values. *, P < 0.05, **, P < 0.01.
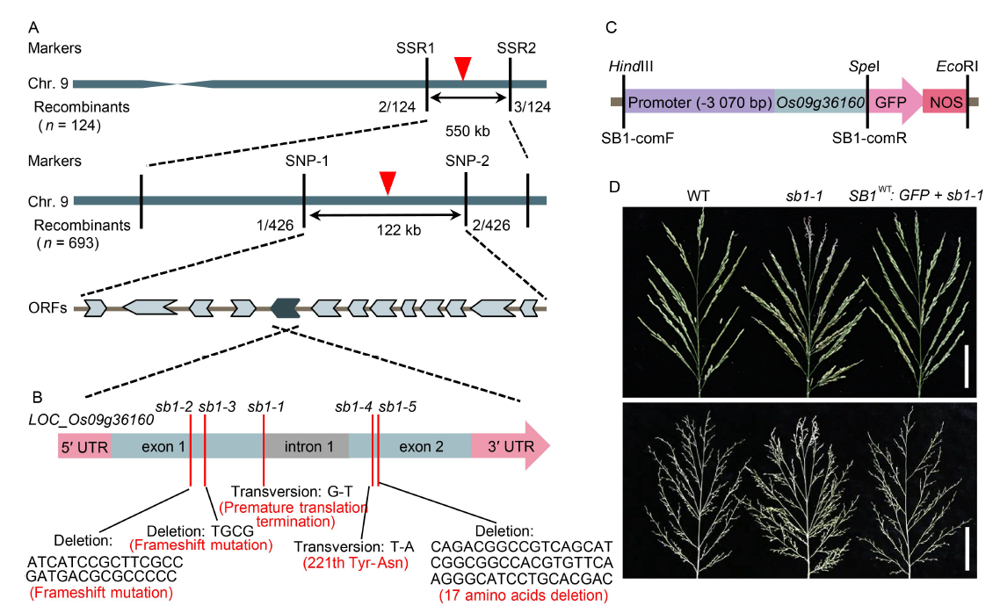
Fig. 2. Map-based cloning of SB1. A, Fine mapping of SB1 on chromosome 9. B, Structure of SB1 and mutation sites of sb1-1/2/3/4/5. C, Structure of the SB1-GFP fusion complementary vector. D, Phenotypes of the wild type, sb1-1 and sb1-1-GFP transgenic plant. Bars are 5 cm.ORF, Open reading frame; SB1-comF and SB1-comR, Cloning primers of SB1 for complementary test.
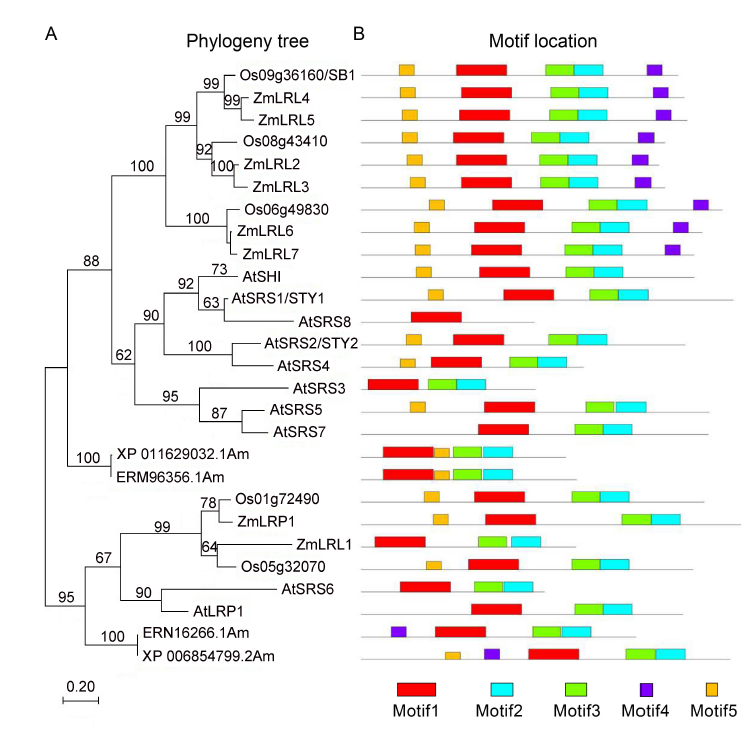
Fig. 3. SB1 encodes a RING-like zinc finger protein.A, Phylogenetic tree analysis. The phylogenetic tree was constructed using the neighbor- joining method based on the Jone-Taylor- Thornton matrix-based model, and bootstrap support values calculated from 1 000 replicates are given at the branch nodes. At, Arabidopisis thaliana; Os, Oryza sativa; Zm, Zea mays. B, Location of conserved domains of the genes in the phylogeny tree.
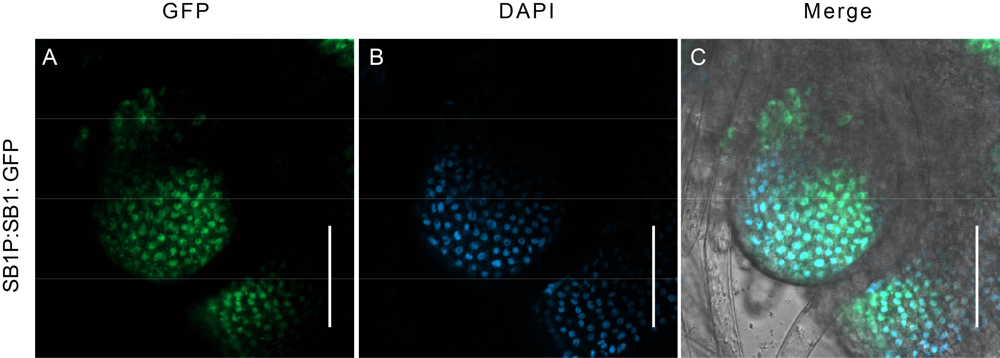
Fig. S4. Subcellular location of SB1 by observing the GFP signal in SB1P:SB1: GFP transgenic plants. A, GFP signals in cells of branch meristem. B, DAPI signal in nucleus. C, Merged fluorescence signal of GFP and DAPI.Bars are 50 μm.
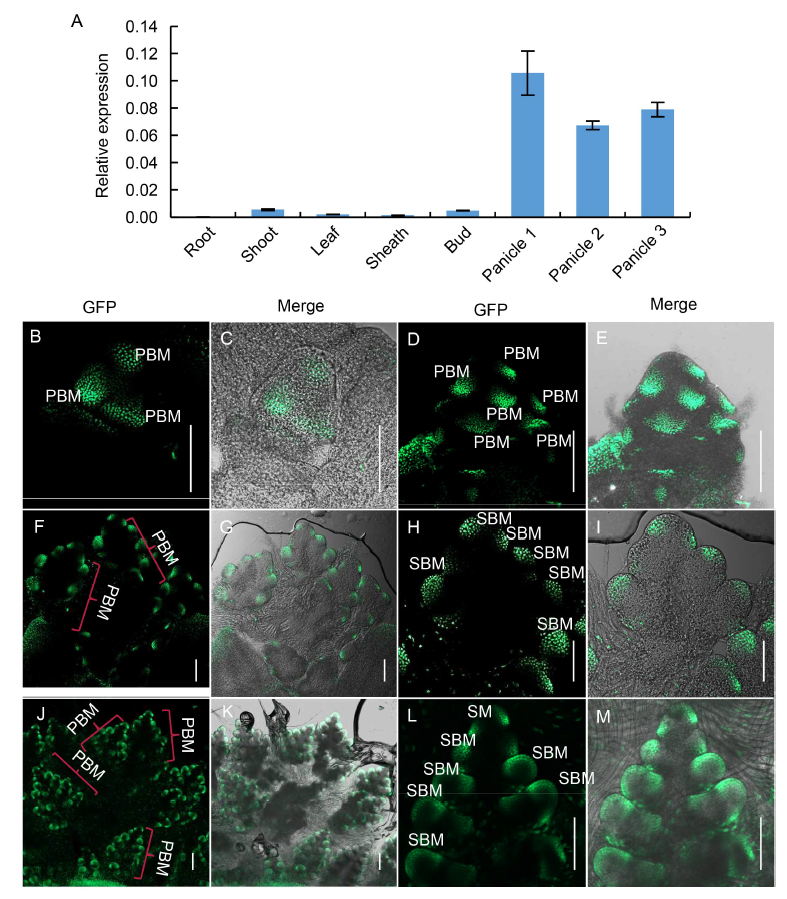
Fig. 4. Expression pattern of SB1. A, qPCR analysis of SB1 in root, shoot, leaf, sheath, tiller bud and panicles of different length. Panicles 1, 2 and 3 refer to panicles with the lengths of < 0.5, 0.5?1.0 and 1.1?2.0 cm, respectively. OsActin was used as a reference gene. Data are Mean ± SD (n = 3).B?M, GFP signal indicating SB1 expression in complementary transgenic plants harboring the construct SB1P: SB1:GFP. B and C, Panicles at the stage In2; D and E, Panicles at the stage In3; F and G, Panicles at the stage In4; H and I were the magnification figure of a PBM in F and G; J and K, Panicles at the stage In5; L and M were the magnification figure of a PBM in J and K. PBM, Primary branch meristem; SBM, Secondary branch meristem; SM, Spikelet meristem. In2, 3, 4 and 5 refer to the four inflorescence stages of rice during which the branch meristems formed. Bars are 100 μm.
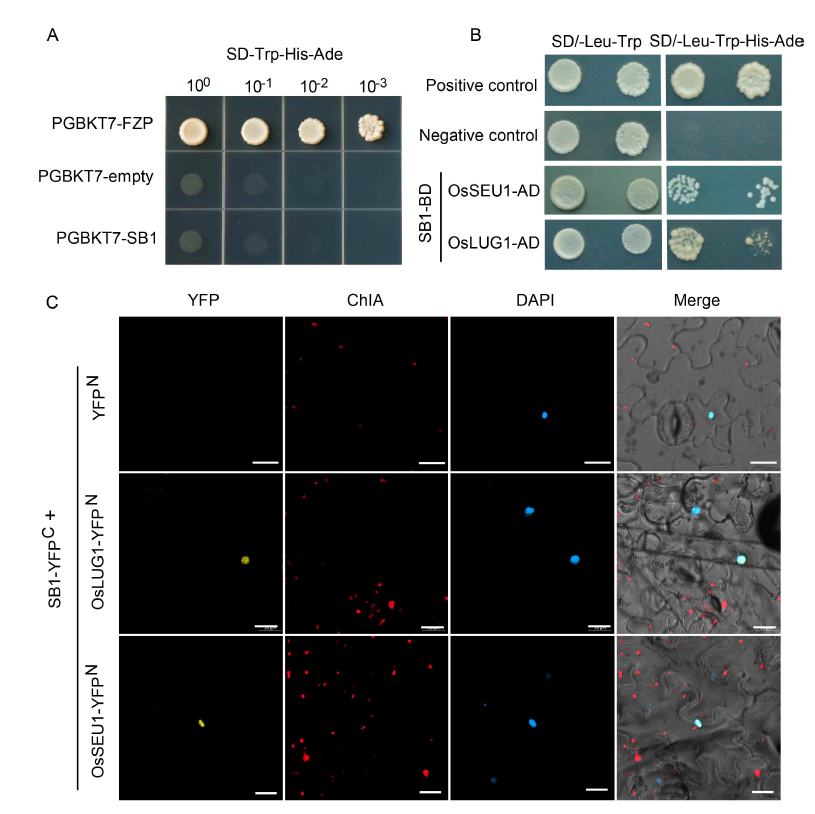
Fig. 5. Interaction of SB1 with corepressors of OsSEU1 and OsLUG1. A, Autoactivation analysis of SB1 protein. PGBKT7-FZP was used as a positive control and PGBKT7-empty as a negative control. B, SB1 interacted with OsSEU1 and OsLUG1 in yeast cells by a yeast two- hybrid assay. C, SB1 interacted with OsSEU1 and OsLUG1 in epidermal cells of Nicotiana benthamiana by a bimolecular fluorescent complimentary (BiFC) assay. YFP, Yellow fluorescent protein; ChlA, Chlorophyll A; DAPI, 4’,6-diamidino-2-phenylindole. Bars are 20 μm.
| No. | ID | Description |
|---|---|---|
| 1 | Os09g0552800 | No apical meristem protein, putative |
| 2 | Os12g0580700 | Zinc finger, C3HC4 type domain containing protein |
| 3 | Os05g0103500 | CHCH domain containing protein |
| 4 | Os03g0685600 | WD40 repeat-like domain containing protein |
| 5 | Os01g0231000 | OsIAA3 - Auxin-responsive Aux/IAA gene family member |
| 6 | Os02g0757100 | Cell cycle gene |
| 7 | Os09g0567000 | DUF1618 domain containing protein |
| 8 | Os02g0170500 | Nuclear transcription factor Y subunit C |
| 9 | Os02g0261100 | Licitor-responsive ubiquitin-conjugating enzyme |
| 10 | Os04g0510200 | Transcriptional corepressor LEUNIG |
| 11 | Os06g0160400 | Heading and grain weight regulat gene |
| 12 | Os02g0203000 | bZIP transcription factor domain containing protein |
| 13 | Os04g0548000 | Ethylene-responsive element-binding protein, putative |
| 14 | Os06g0142600 | Heading date gene, |
| 15 | Os03g0177400 | Elongation factor Tu |
| 16 | Os03g0192400 | NADH dehydrogenase 1 alpha subcomplex subunit 13 |
| 17 | Os02g0255500 | Pyrabactin resistance-like abscisic acid receptor |
| 18 | Os04g0386900 | Transcriptional factor B3 family protein |
| 19 | Os10g0575900 | Expressed protein |
| 20 | Os05g0411300 | bZIP transcription factor |
| 21 | Os04g0550600 | Carotenoid cleavage dioxygenase |
| 22 | Os04g0615000 | Narrow leaf NAL1, |
| 23 | Os05g0573500 | Nuclear transcription factor Y subunit B |
| 24 | Os07g0600400 | WD repeat-containing protein 12 |
| 25 | Os06g0126000 | Transcriptional corepressor SEUSS |
| 26 | Os02g0170500 | Nuclear transcription factor Y subunit C |
| 27 | Os09g0369050 | Similar to DRE binding factor 2 |
| 28 | Os03g0775500 | Expressed protein |
| 29 | Os02g0723700 | SWI/SNF-related regulator |
| 30 | Os02g0122000 | Histone deacetylase complex subunit SAP18 |
| 31 | Os03g0122100 | Helix-loop-helix DNA-binding domain containing protein |
Table S1. Interacting proteins of SB1 identified by yeast two-hybrid screening.
| No. | ID | Description |
|---|---|---|
| 1 | Os09g0552800 | No apical meristem protein, putative |
| 2 | Os12g0580700 | Zinc finger, C3HC4 type domain containing protein |
| 3 | Os05g0103500 | CHCH domain containing protein |
| 4 | Os03g0685600 | WD40 repeat-like domain containing protein |
| 5 | Os01g0231000 | OsIAA3 - Auxin-responsive Aux/IAA gene family member |
| 6 | Os02g0757100 | Cell cycle gene |
| 7 | Os09g0567000 | DUF1618 domain containing protein |
| 8 | Os02g0170500 | Nuclear transcription factor Y subunit C |
| 9 | Os02g0261100 | Licitor-responsive ubiquitin-conjugating enzyme |
| 10 | Os04g0510200 | Transcriptional corepressor LEUNIG |
| 11 | Os06g0160400 | Heading and grain weight regulat gene |
| 12 | Os02g0203000 | bZIP transcription factor domain containing protein |
| 13 | Os04g0548000 | Ethylene-responsive element-binding protein, putative |
| 14 | Os06g0142600 | Heading date gene, |
| 15 | Os03g0177400 | Elongation factor Tu |
| 16 | Os03g0192400 | NADH dehydrogenase 1 alpha subcomplex subunit 13 |
| 17 | Os02g0255500 | Pyrabactin resistance-like abscisic acid receptor |
| 18 | Os04g0386900 | Transcriptional factor B3 family protein |
| 19 | Os10g0575900 | Expressed protein |
| 20 | Os05g0411300 | bZIP transcription factor |
| 21 | Os04g0550600 | Carotenoid cleavage dioxygenase |
| 22 | Os04g0615000 | Narrow leaf NAL1, |
| 23 | Os05g0573500 | Nuclear transcription factor Y subunit B |
| 24 | Os07g0600400 | WD repeat-containing protein 12 |
| 25 | Os06g0126000 | Transcriptional corepressor SEUSS |
| 26 | Os02g0170500 | Nuclear transcription factor Y subunit C |
| 27 | Os09g0369050 | Similar to DRE binding factor 2 |
| 28 | Os03g0775500 | Expressed protein |
| 29 | Os02g0723700 | SWI/SNF-related regulator |
| 30 | Os02g0122000 | Histone deacetylase complex subunit SAP18 |
| 31 | Os03g0122100 | Helix-loop-helix DNA-binding domain containing protein |
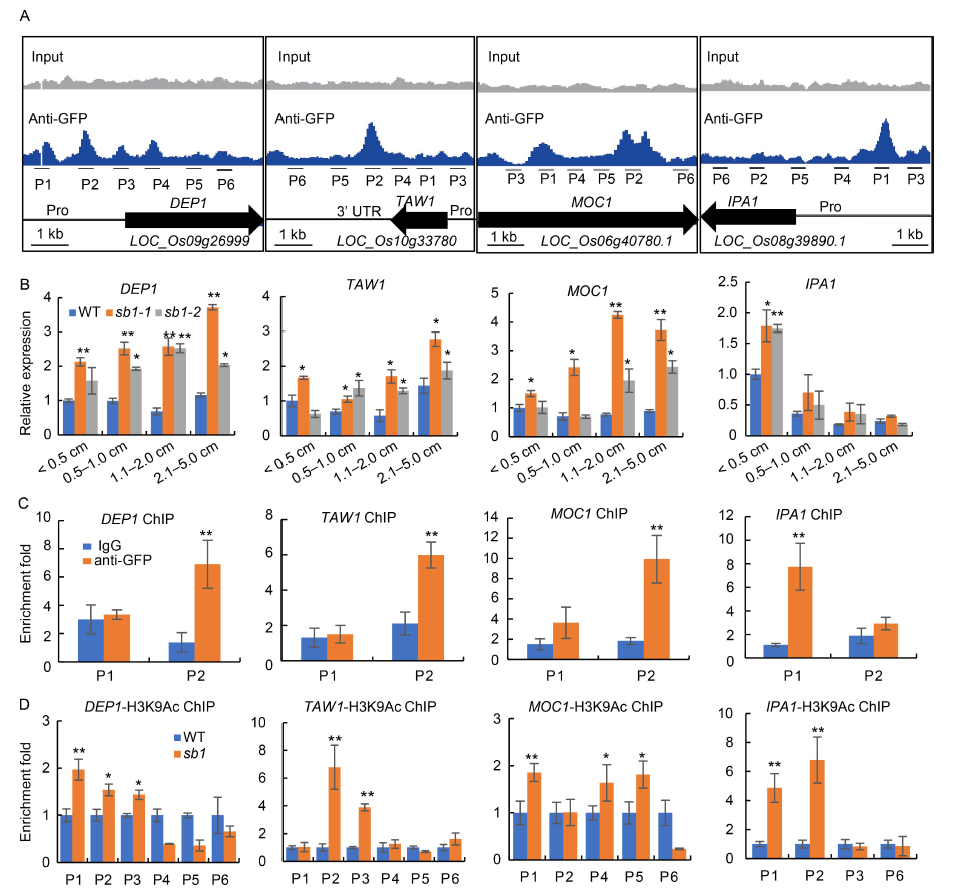
Fig. 6. SB1 directly repressed expression of DEP1, TAW1, MOC1 and IPA1. A, Binding peaks of SB1 with DEP1, TAW1, MOC1 and IPA1 by chromatin immunoprecipitation-sequence (ChIP-seq) using an anti-GFP antibody.B, Relative expression of DEP1, TAW1, MOC1 and IPA1 in the wild type (WT) and sb1 panicles, respectively.C, ChIP-qPCR for P1 and P2 sites of DEP1, TAW1, MOC1 and IPA1 with anti-GFP antibody, respectively. ChIP enrichment compared with the input sample was tested by qPCR. D, ChIP-qPCR for several sites of DEP1, TAW1, MOC1 and IPA1 with a H3K9Ac antibody, respectively. ChIP enrichment compared with the IgG sample was tested by qPCR.Pro, Promoter; P1 to P6 represent primers for ChIP-qPCR. Error bars indicated SD of three repeats. The Student’s t-test was used to generate the P values. *, P < 0.05; **, P < 0.01.
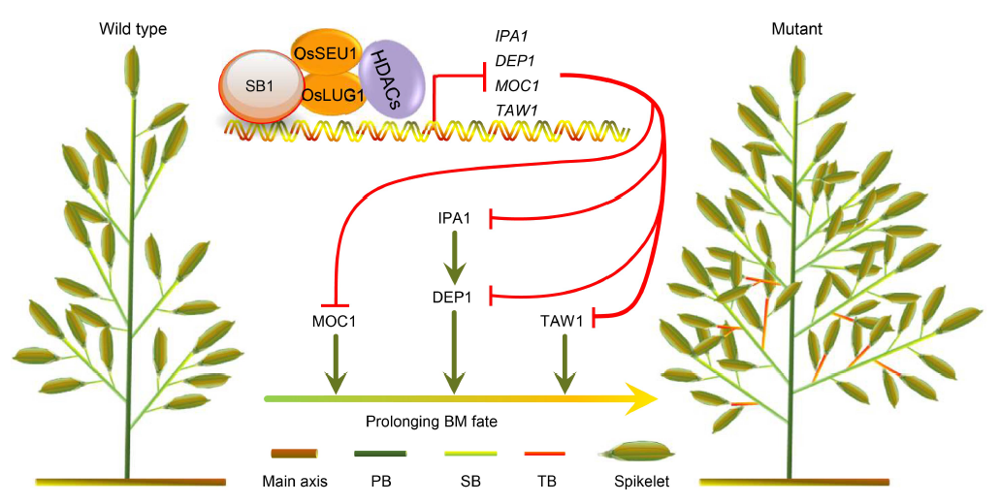
Fig. 7. Model of SB1 gene regulating rice panicle architecture. In the wild type plants, SB1 interacts with the corepressors OsSEU1 and OsLUG1 to form the repression complex and repress DEP1, TAW1, MOC1 and IPA1 by downregulating the acetylation levels of histone on chromosome. In the sb1-1/2/3/4/5 mutants, the repression complex cannot work, resulting in the increased acetylation levels and the expression levels of DEP1, TAW1, MOC1 and IPA1, which enables the formation of the increased numbers of branches and spikelets.HDACs, Histone deacetylases; BM, Branch meristem; PB, Primary branch; SB, Secondary branch; TB, Tertiary branch.
| Purpose | Primer name | Sequence |
|---|---|---|
| Mapping | SSR1-F | CAGTGGCGTGGAGAGAAATTTGG |
| SSR1-R | CTCACCTGCGACAGCAAGATCG | |
| SSR2-F | TGAGCACCATGCAATAACTGTCG | |
| SSR2-R | ACCATCTATCGCCACATCTCACC | |
| SNP-1-F | CGGGACATTCTTAGCTTTGCTTTGAT | |
| SNP-1-R | GAAAGTAGTAGTCTCACCATGCTTTGCCAC | |
| SNP-2-F | CCCACACTGACTGGGACTCACTCG | |
| SNP-2-R | GATTAGCGTCTCGTCCTCGCAGC | |
| Candidate gene cloning | SB1-F | GCAATGGCACAAGTGACACTACAAGA |
| SB1-R | CTCCGATCAGTTCACGCAAAGTCG | |
| Complementation test | SB1-com-F | GTATTTGTCTGTGCTGGATCCGTGTGCGAAGGGATACATGATGAAGC |
| SB1-com-R | CGCCCTTGCTCACCATACTAGTCGGCCGCGGGTGGCC | |
| qPCR | Actin-F | TGCTATGTACGTCGCCATCCAG |
| Actin-R | AATGAGTAACCACGCTCCGTCA | |
| RT-SB1-F | AACGCCACTACAATCTCTCACTCGC | |
| RT-SB1-R | AGTCTAAAGGGTTCGCCTGCCTAA | |
| RT- MOC1-F | CTGTGGTTGCATGACACAGATGCAC | |
| RT- MOC1-R | CCTTGGATGGAATGTCTCAGTCCC | |
| RT- DEP1-F | CCGTTTCTCGTTCTGGAT | |
| RT- DEP1-R | ATCTGTGCCTCCTTCTCT | |
| RT-IPA1-F | TCTTCTGTCAACCCAGCCAT | |
| RT-IPA1-R | GCCATCAAAGCTGGTGGTAG | |
| RT-TAW1-F | GACTGGAACACGTTCGGGCAGTAC | |
| RT-TAW1-R | CACCTTGGTCTTGCCGAACTGGT | |
| RT-LAX1-F | TGACGACGCTGGAGATGGCG | |
| RT-LAX1-R | GGAGGCAGCTGATGAGCGCC | |
| RT-RCN11-F | GACAGCAACCTGCTCAAGGTCATCTC | |
| RT-RCN1-R | GCTGGTCGATTAGCTCCTTCACCC | |
| RT-OSH1-F | CAGGACCTGGAGCTTCGCCAG | |
| RT-OSH1-R | GGAGAGCGTGTTGAGCTGCGTCT | |
| RT-MOC3-F | GGATCAGGCCAACGTCGTCAACT | |
| RT-MOC3-R | CGACTGGGAAGAGTGGAAGCGTCT | |
| Two-Hybrid System | SB1-BD-F1 | ATGGGCAGCGGCGGCGGT |
| SB1-BD-R1 | GAACATCCCACGGTTTGGAGGTT | |
| SEU-AD-F1 | ATGTCCAACCTCCAAACCGTGGG | |
| SEU-AD-R1- | CTAAGATCCACAGATGGACATGGAGAG | |
| LEU-AD-F1 | ATGTCGTCGCTCAGCCGGGAG | |
| LEU-AD-R1 | CTATCTTTGGATTTGGTCTGCAGCT | |
| BiFC assay | SB1-YC-F | CAATTACAGGTACCCGGGGATCCATGGCGGGGTTCCCTCTAGGC |
| SB1-YC-R | TGCACGCTGCCACCGCCGTCGACCGGCCGCGGGTGGCCGTGGAA | |
| OsLUG1-YN-F | CATCGAGGACGCCGGCGGATCCATGGCGCAGAGCAACTGGGAAG | |
| OsLUG1-YN-R | GAACGAAAGCTCTGCAGGTCGACCTTCCACAGCTTGACAGAGTTGTCG | |
| OsSEU1-YN-F | CATCGAGGACGCCGGCGGATCCATGTCTGGGGCGCCATGCTC | |
| OsSEU1-YN-R | GAACGAAAGCTCTGCAGGTCGACCATGTTCCATGAGTAGCCACCACCT | |
| ChIP-qPCR | DEP1-P1-F | CGCCCACACACAACACAGCTAG |
| DEP1-P1-R | GACCGGGAACACACGTTGTCA | |
| DEP1-P2-F | AGCTCAACTGAACGCTGGCTG | |
| DEP1-P2-R | CCACCACTGCTGCTACTGCCTA | |
| DEP1-P3-F | ACGAAGGATCGGCTTTGCAT | |
| DEP1-P3-R | CGAGCCTACGTTGGATACGCT | |
| DEP1-P4-F | GGAGCTACCCGCTACTGCAAG | |
| DEP1-P4-R | CGTGATTCACCTCCGCCTAGA | |
| DEP1-P5-F | CCTGCATCATAACGTTCCTAGTGGT | |
| DEP1-P5-R | CTCAGGACTGTGAGCTGTAATATGG | |
| DEP1-P6-F | GCTGTAGTCCAGACTGCTGCTCATG | |
| DEP1-P6-R | GTTGCACTGGGACTTGAAGCATG | |
| TAW1-P1-F | TGGACCAGTTCGGCAAGACCAAG | |
| TAW1-P1-R | TGCTCGCGGACCTCGCGGAGGTAG | |
| TAW1-P2-F | TCGAGTCATGTCAGCACTGTGGC | |
| TAW1-P2-R | GCATCACTCTCTTAACGCTGCTGC | |
| TAW1-P3-F | TCTCTCTCTCTCTTGGACCACTGCG | |
| TAW1-P3-R | CGAACTCCATCGTCGATCTGCAC | |
| TAW1-P4-F | CGAGCAGGTACGAGTCGCAGAAG | |
| TAW1-P4-R | CTTGGTCTTGCCGAACTGGTCC | |
| TAW1-P5-F | ACTCCACTCCACTCCACTCCACTATG | |
| TAW1-P5-R | TAGTAGACGAGACGACGTGCGAGAG | |
| TAW1-P6-F | CTCTCTCTCTGCTGCTGCAACAAG | |
| TAW1-P6-R | CTGCCTGACTGACTGACTCTCTCTG | |
| MOC1-P1-F | GAGTGGCACTTGCTACTGTGCATG | |
| MOC1-P1-R | TGATGATGGCCTCCAGATAACTTCC | |
| MOC1-P2-F | TATGGCACAAACAGTCACAGCTGC | |
| MOC1-P2-R | TGCTCATTCCTTGATGAGAGAGTGAG | |
| MOC1-P4-F | GTGGAGAGCCGGAATCAACACTGT | |
| MOC1-P4-R | GACATATAATGATACTCCGGCCATCTC | |
| MOC1-P5-F | GCTGGGATTGCACAGTGAAGCTAG | |
| MOC1-P5-R | GCACGTCCAACATCTGGGCTT | |
| MOC1-P6-F | TACCTGGCGTTCAACCAGATCG | |
| MOC1-P6-R | CGTCGAGGTCGAGGATGTGGAC | |
| IPA1-P1-F | GAGTGGCACTTGCTACTGTGCATG | |
| IPA1-P1-R | TGATGATGGCCTCCAGATAACTTCC | |
| IPA1-P2-F | TATGGCACAAACAGTCACAGCTGC | |
| IPA1-P2-R | TGCTCATTCCTTGATGAGAGAGTGAG | |
| IPA1-P3-F | CCAGTTACAACCCTCCACCATTCAC | |
| IPA1-P3-R | TGTGGCAGTGCCCACAGTGTGT | |
| IPA1-P6-F | GACGTGGCGTGTGGCAATGTAG | |
| IPA1-P6-R | TACTCCGACGAGCCTCCTTTCTCT |
Table S2. Primers used in the study.
| Purpose | Primer name | Sequence |
|---|---|---|
| Mapping | SSR1-F | CAGTGGCGTGGAGAGAAATTTGG |
| SSR1-R | CTCACCTGCGACAGCAAGATCG | |
| SSR2-F | TGAGCACCATGCAATAACTGTCG | |
| SSR2-R | ACCATCTATCGCCACATCTCACC | |
| SNP-1-F | CGGGACATTCTTAGCTTTGCTTTGAT | |
| SNP-1-R | GAAAGTAGTAGTCTCACCATGCTTTGCCAC | |
| SNP-2-F | CCCACACTGACTGGGACTCACTCG | |
| SNP-2-R | GATTAGCGTCTCGTCCTCGCAGC | |
| Candidate gene cloning | SB1-F | GCAATGGCACAAGTGACACTACAAGA |
| SB1-R | CTCCGATCAGTTCACGCAAAGTCG | |
| Complementation test | SB1-com-F | GTATTTGTCTGTGCTGGATCCGTGTGCGAAGGGATACATGATGAAGC |
| SB1-com-R | CGCCCTTGCTCACCATACTAGTCGGCCGCGGGTGGCC | |
| qPCR | Actin-F | TGCTATGTACGTCGCCATCCAG |
| Actin-R | AATGAGTAACCACGCTCCGTCA | |
| RT-SB1-F | AACGCCACTACAATCTCTCACTCGC | |
| RT-SB1-R | AGTCTAAAGGGTTCGCCTGCCTAA | |
| RT- MOC1-F | CTGTGGTTGCATGACACAGATGCAC | |
| RT- MOC1-R | CCTTGGATGGAATGTCTCAGTCCC | |
| RT- DEP1-F | CCGTTTCTCGTTCTGGAT | |
| RT- DEP1-R | ATCTGTGCCTCCTTCTCT | |
| RT-IPA1-F | TCTTCTGTCAACCCAGCCAT | |
| RT-IPA1-R | GCCATCAAAGCTGGTGGTAG | |
| RT-TAW1-F | GACTGGAACACGTTCGGGCAGTAC | |
| RT-TAW1-R | CACCTTGGTCTTGCCGAACTGGT | |
| RT-LAX1-F | TGACGACGCTGGAGATGGCG | |
| RT-LAX1-R | GGAGGCAGCTGATGAGCGCC | |
| RT-RCN11-F | GACAGCAACCTGCTCAAGGTCATCTC | |
| RT-RCN1-R | GCTGGTCGATTAGCTCCTTCACCC | |
| RT-OSH1-F | CAGGACCTGGAGCTTCGCCAG | |
| RT-OSH1-R | GGAGAGCGTGTTGAGCTGCGTCT | |
| RT-MOC3-F | GGATCAGGCCAACGTCGTCAACT | |
| RT-MOC3-R | CGACTGGGAAGAGTGGAAGCGTCT | |
| Two-Hybrid System | SB1-BD-F1 | ATGGGCAGCGGCGGCGGT |
| SB1-BD-R1 | GAACATCCCACGGTTTGGAGGTT | |
| SEU-AD-F1 | ATGTCCAACCTCCAAACCGTGGG | |
| SEU-AD-R1- | CTAAGATCCACAGATGGACATGGAGAG | |
| LEU-AD-F1 | ATGTCGTCGCTCAGCCGGGAG | |
| LEU-AD-R1 | CTATCTTTGGATTTGGTCTGCAGCT | |
| BiFC assay | SB1-YC-F | CAATTACAGGTACCCGGGGATCCATGGCGGGGTTCCCTCTAGGC |
| SB1-YC-R | TGCACGCTGCCACCGCCGTCGACCGGCCGCGGGTGGCCGTGGAA | |
| OsLUG1-YN-F | CATCGAGGACGCCGGCGGATCCATGGCGCAGAGCAACTGGGAAG | |
| OsLUG1-YN-R | GAACGAAAGCTCTGCAGGTCGACCTTCCACAGCTTGACAGAGTTGTCG | |
| OsSEU1-YN-F | CATCGAGGACGCCGGCGGATCCATGTCTGGGGCGCCATGCTC | |
| OsSEU1-YN-R | GAACGAAAGCTCTGCAGGTCGACCATGTTCCATGAGTAGCCACCACCT | |
| ChIP-qPCR | DEP1-P1-F | CGCCCACACACAACACAGCTAG |
| DEP1-P1-R | GACCGGGAACACACGTTGTCA | |
| DEP1-P2-F | AGCTCAACTGAACGCTGGCTG | |
| DEP1-P2-R | CCACCACTGCTGCTACTGCCTA | |
| DEP1-P3-F | ACGAAGGATCGGCTTTGCAT | |
| DEP1-P3-R | CGAGCCTACGTTGGATACGCT | |
| DEP1-P4-F | GGAGCTACCCGCTACTGCAAG | |
| DEP1-P4-R | CGTGATTCACCTCCGCCTAGA | |
| DEP1-P5-F | CCTGCATCATAACGTTCCTAGTGGT | |
| DEP1-P5-R | CTCAGGACTGTGAGCTGTAATATGG | |
| DEP1-P6-F | GCTGTAGTCCAGACTGCTGCTCATG | |
| DEP1-P6-R | GTTGCACTGGGACTTGAAGCATG | |
| TAW1-P1-F | TGGACCAGTTCGGCAAGACCAAG | |
| TAW1-P1-R | TGCTCGCGGACCTCGCGGAGGTAG | |
| TAW1-P2-F | TCGAGTCATGTCAGCACTGTGGC | |
| TAW1-P2-R | GCATCACTCTCTTAACGCTGCTGC | |
| TAW1-P3-F | TCTCTCTCTCTCTTGGACCACTGCG | |
| TAW1-P3-R | CGAACTCCATCGTCGATCTGCAC | |
| TAW1-P4-F | CGAGCAGGTACGAGTCGCAGAAG | |
| TAW1-P4-R | CTTGGTCTTGCCGAACTGGTCC | |
| TAW1-P5-F | ACTCCACTCCACTCCACTCCACTATG | |
| TAW1-P5-R | TAGTAGACGAGACGACGTGCGAGAG | |
| TAW1-P6-F | CTCTCTCTCTGCTGCTGCAACAAG | |
| TAW1-P6-R | CTGCCTGACTGACTGACTCTCTCTG | |
| MOC1-P1-F | GAGTGGCACTTGCTACTGTGCATG | |
| MOC1-P1-R | TGATGATGGCCTCCAGATAACTTCC | |
| MOC1-P2-F | TATGGCACAAACAGTCACAGCTGC | |
| MOC1-P2-R | TGCTCATTCCTTGATGAGAGAGTGAG | |
| MOC1-P4-F | GTGGAGAGCCGGAATCAACACTGT | |
| MOC1-P4-R | GACATATAATGATACTCCGGCCATCTC | |
| MOC1-P5-F | GCTGGGATTGCACAGTGAAGCTAG | |
| MOC1-P5-R | GCACGTCCAACATCTGGGCTT | |
| MOC1-P6-F | TACCTGGCGTTCAACCAGATCG | |
| MOC1-P6-R | CGTCGAGGTCGAGGATGTGGAC | |
| IPA1-P1-F | GAGTGGCACTTGCTACTGTGCATG | |
| IPA1-P1-R | TGATGATGGCCTCCAGATAACTTCC | |
| IPA1-P2-F | TATGGCACAAACAGTCACAGCTGC | |
| IPA1-P2-R | TGCTCATTCCTTGATGAGAGAGTGAG | |
| IPA1-P3-F | CCAGTTACAACCCTCCACCATTCAC | |
| IPA1-P3-R | TGTGGCAGTGCCCACAGTGTGT | |
| IPA1-P6-F | GACGTGGCGTGTGGCAATGTAG | |
| IPA1-P6-R | TACTCCGACGAGCCTCCTTTCTCT |
| [1] | Ashikari M, Sakakibara H, Lin S Y, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production. Science, 309: 741‒745. |
| [2] | Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. 1997. Inflorescence commitment and architecture in Arabidopsis. Science, 275: 80‒83. |
| [3] | Conner J, Liu Z. 2000. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA, 97(23): 12902‒12907. |
| [4] | Duan E C, Wang Y H, Li X H, Lin Q B, Zhang T, Wang Y P, Zhou C L, Zhang H, Jiang L, Wang J L, Lei C L, Zhang X, Guo X P, Wang H Y, Wan J M. 2019. OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell, 31(5): 1026. |
| [5] | Franks R G, Wang C X, Levin J Z, Liu Z C. 2002. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development, 129(1): 253‒263. |
| [6] | Fridborg I, Kuusk S, Robertson M, Sundberg E. 2001. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol, 127(3): 937‒948. |
| [7] | Gao X C, Liang W Q, Yin C S, Ji S M, Wang H M, Su X, Guo C C, Kong H Z, Xue H W, Zhang D B. 2010. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol, 153(2): 728‒740. |
| [8] | Gonzalez D, Bowen A J, Carroll T S, Conlan R S. 2007. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol, 27(15): 5306‒5315. |
| [9] | Guo S Y, Sun B, Looi L S, Xu Y F, Gan E S, Huang J B, Ito T. 2015. Co-ordination of flower development through epigenetic regulation in two model species: Rice and Arabidopsis. Plant Cell Physiol, 56(5): 830‒842. |
| [10] | Hu B, Wang W, Ou S J, Tang J Y, Li H, Che R H, Zhang Z H, Chai X Y, Wang H R, Wang Y Q, Liang C Z, Liu L C, Piao Z Z, Deng Q Y, Deng K, Xu C, Liang Y, Zhang L H, Li L G, Chu C C. 2015. Variation in NRT1. 1B contributes to nitrate-use divergence between rice subspecies. Nat Genet, 47(7): 834‒838. |
| [11] | Huang W T, Bai G X, Wang J, Zhu W, Zeng Q S. 2018. Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front Plant Sci, 9: 300. |
| [12] | Huang X Z, Qian Q, Liu Z B, Sun H Y, He S Y, Luo D, Xia G M, Chu C C, Li J Y, Fu X D. 2009. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet, 41(4): 494‒497. |
| [13] | Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J I, Nagato Y. 2012. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J, 69(1): 168‒180. |
| [14] | Jiao Y Q, Wang Y H, Xue D W, Wang J, Yan M X, Liu G F, Dong G J, Zeng D L, Lu Z F, Zhu X D, Qian Q, Li J Y. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet, 42(6): 541‒544. |
| [15] | Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. 2010. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol, 51(1): 47‒57. |
| [16] | Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature, 445: 652‒655. |
| [17] | Kuusk S, Sohlberg J J, Long J A, Fridborg I, Sundberg E. 2002. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development, 129(20): 4707‒4717. |
| [18] | Kuusk S, Sohlberg J J, Eklund D M, Sundberg E. 2010. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J, 47(1): 99‒111. |
| [19] | Li F, Liu W B, Tang J Y, Chen J F, Tong H N, Hu B, Li C L, Fang J, Chen M S, Chu C C. 2010. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res, 20(7): 838‒849. |
| [20] | Li M, Tang D, Wang K J, Wu X R, Lu L L, Yu H X, Gu M H, Yan C J, Cheng Z K. 2011. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J, 9(9): 1002‒1013. |
| [21] | Li S B, Qian Q, Fu Z M, Zeng D L, Meng X B, Kyozuka J, Maekawa M, Zhu X D, Zhang J, Li J Y, Wang Y H. 2009. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J, 58(4): 592‒605. |
| [22] | Li S Y, Zhao B R, Yuan D Y, Duan M J, Qian Q, Tang L, Wang B, Liu X Q, Zhang J, Wang J, Sun J Q, Liu Z, Feng Y Q, Yuan L P, Li C Y. 2013. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA, 110(8): 3167‒3172. |
| [23] | Li W Q, Wu J G, Weng S L, Zhang Y J, Zhang D P, Shi C H. 2010. Identification and characterization of dwarf 62, a loss-of- function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta, 232(6): 1383‒1396. |
| [24] | Li X G, Qian Q, Fu Z M, Wang Y H, Xiong G S, Zeng D L, Wang X Q, Liu X F, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li J Y. 2003. Control of tillering in rice. Nature, 422: 618‒621. |
| [25] | Li Z K, Pinson S R M, Stansel J W, Paterson A H. 1998. Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (Oryza sativa L.). Mol Breeding, 4(5): 419‒426. |
| [26] | Liu Z C, Karmarkar V. 2008. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci, 13(3): 137‒144. |
| [27] | Lu Z F, Yu H, Xiong G S, Wang J, Jiao Y Q, Liu G F, Jing Y H, Meng X B, Hu X M, Qian Q, Fu X D, Wang Y H, Li J Y. 2013. Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell, 25(10): 3743‒3759. |
| [28] | Mimida N, Goto K, Kobayashi Y, Araki T, Ahn J H, Weigel D, Murata M, Motoyoshi F, Sakamoto W. 2001. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells, 6(4): 327‒336. |
| [29] | Miura K, Ikeda M, Matsubara A, Song X J, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet, 42(6): 545‒549. |
| [30] | Nakagawa M, Shimamoto K, Kyozuka J. 2002. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J, 29(6): 743‒750. |
| [31] | Rao N N, Prasad K, Kumar P R, Vijayraghavan U. 2008. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA, 105(9): 3646‒3651. |
| [32] | Ratcliffe O J, Bradley D J, Coen E S. 1999. Separation of shoot and floral identity in Arabidopsis. Development, 126(6): 1109‒1120. |
| [33] | Sheng P K, Wu F Q, Tan J J, Zhang H, Ma W W, Chen L P, Wang J C, Wang J, Zhu S S, Guo X P, Wang J L, Zhang X, Cheng Z J, Bao Y Q, Wu C Y, Liu X M, Wan J M. 2016. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol Biol, 92(1): 209‒222. |
| [34] | Shi B H, Zhang C, Tian C H, Wang J, Wang Q, Xu T F, Xu Y, Ohno C, Sablowski R, Heisler M G, Theres K, Wang Y, Jiao Y L. 2016. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet, 12(7): e1006168. |
| [35] | Sitaraman J, Bui M, Liu Z C. 2008. LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol, 147(2): 672‒681. |
| [36] | Skirpan A, Wu X T, McSteen P. 2008. Genetic and physical interaction suggest that BARREN STALK1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J, 55(5): 787‒797. |
| [37] | Sridhar V V, Surendrarao A, Gonzalez D, Steven Conlan R, Liu Z C. 2004. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA, 101(31): 11494‒11499. |
| [38] | Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang X H, Yoshida H, Kyozuka J, Chen F, Sato Y. 2011. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell, 23(9): 3276‒3287. |
| [39] | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol, 28(10): 2731‒2739. |
| [40] | Teng S, Qian Q, Zeng D L, Kunihiro Y, Huang D N, Zhu L H. 2002. QTL analysis of rice peduncle vascular bundle system and panicle traits. Acta Bot Sin, 44(3): 301‒306. (in Chinese with English abstract) |
| [41] | Wang L, Zeng X Q, Zhuang H, Shen Y L, Chen H, Wang Z W, Long J C, Ling Y H, He G H, Li Y F. 2017. Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice. Plant Growth Regul, 81(3): 433‒442. |
| [42] | Wang W, Hu B, Yuan D Y, Liu Y Q, Che R H, Hu Y C, Ou S J, Liu Y X, Zhang Z H, Wang H R, Li H, Jiang Z M, Zhang Z L, Gao X K, Qiu Y H, Meng X B, Liu Y X, Bai Y, Liang Y, Wang Y Q, Zhang L H, Li L G, Sodmergen S, Jing H C, Li J Y, Chu C C. 2018. Expression of the nitrate transporter gene OsNRT1.1A/ OsNPF6.3 confers high yield and early maturation in rice. Plant Cell, 30(3): 638‒651. |
| [43] | Wang Y H, Li J Y. 2011. Branching in rice. Curr Opin Plant Biol, 14(1): 94‒99. |
| [44] | Wei X J, Xu J F, Guo H N, Jiang L, Chen S H, Yu C, Zhou Z L, Hu P S, Zhai H Q, Wan J M. 2010. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol, 153(4): 1747‒1758. |
| [45] | Wu Y, Wang Y, Mi X F, Shan J X, Li X M, Xu J L, Lin H X. 2016. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet, 12(10): e1006386. |
| [46] | Xiao H, Tang J F, Li Y F, Wang W M, Li X B, Jin L, Xie R, Luo H F, Zhao X F, Meng Z, He G H, Zhu L H. 2009. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J, 59(5): 789‒801. |
| [47] | Xue W Y, Xing Y Z, Weng X Y, Zhao Y, Tang W J, Wang L, Zhou H J, Yu S B, Xu C G, Li X H, Zhang Q F. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet, 40(6): 761‒767. |
| [48] | Yan W H, Wang P, Chen H X, Zhou H J, Li Q P, Wang C R, Ding Z H, Zhang Y S, Yu S B, Xing Y Z, Zhang Q F. 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant, 4(2): 319‒330. |
| [49] | Yang J, Cho L H, Yoon J, Yoon H, Wai A H, Hong W J, Han M, Sakakibara H, Liang W Q, Jung K H, Jeon J S, Koh H J, Zhang D B, An G. 2019. Chromatin interacting factor OsVIL2 increases biomass and rice grain yield. Plant Biotechnol J, 17(1): 178‒187. |
| [50] | Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R H, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, Nagamura Y, Ushijima T, Kumamaru T, Iida S, Maekawa M, Kyozuka J. 2013. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA, 110(2): 767‒772. |
| [51] | Zhang T, Li Y F, Ma L, Sang X C, Ling Y H, Wang Y T, Yu P, Zhuang H, Huang J Y, Wang N, Zhao F M, Zhang C W, Yang Z L, Fang L K, He G H. 2017. LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc Natl Acad Sci USA, 114: 9984‒9989. |
| [52] | Zhuang H, Wang H L, Zhang T, Zeng X Q, Chen H, Wang Z W, Zhang J, Zheng H, Tang J, Ling Y H, Yang Z L, He G H, Li Y F. 2020. NONSTOP GLUMES1 encodes a C2H2 zinc finger protein that regulates spikelet development in rice. Plant Cell, 32(2): 392‒413. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||