
Rice Science ›› 2021, Vol. 28 ›› Issue (5): 479-792.DOI: 10.1016/j.rsci.2021.07.008
• Research Paper • Previous Articles Next Articles
Saichompoo Uthomphon1, Narumol Possawat1, Nakwilai Pawat1, Thongyos Peeranut1, Nanta Aekchupong2, Tippunya Patompong2, Ruengphayak Siriphat3, Itthisoponkul Teerarat4, Bueraheng Niranee5, Cheabu Sulaiman6, Malumpong Chanate1( )
)
Received:2020-08-10
Accepted:2020-10-26
Online:2021-09-28
Published:2021-09-28
Saichompoo Uthomphon, Narumol Possawat, Nakwilai Pawat, Thongyos Peeranut, Nanta Aekchupong, Tippunya Patompong, Ruengphayak Siriphat, Itthisoponkul Teerarat, Bueraheng Niranee, Cheabu Sulaiman, Malumpong Chanate. Breeding Novel Short Grain Rice for Tropical Region to Combine Important Agronomical Traits, Biotic Stress Resistance and Cooking Quality in Koshihikari Background[J]. Rice Science, 2021, 28(5): 479-792.
Add to citation manager EndNote|Ris|BibTeX
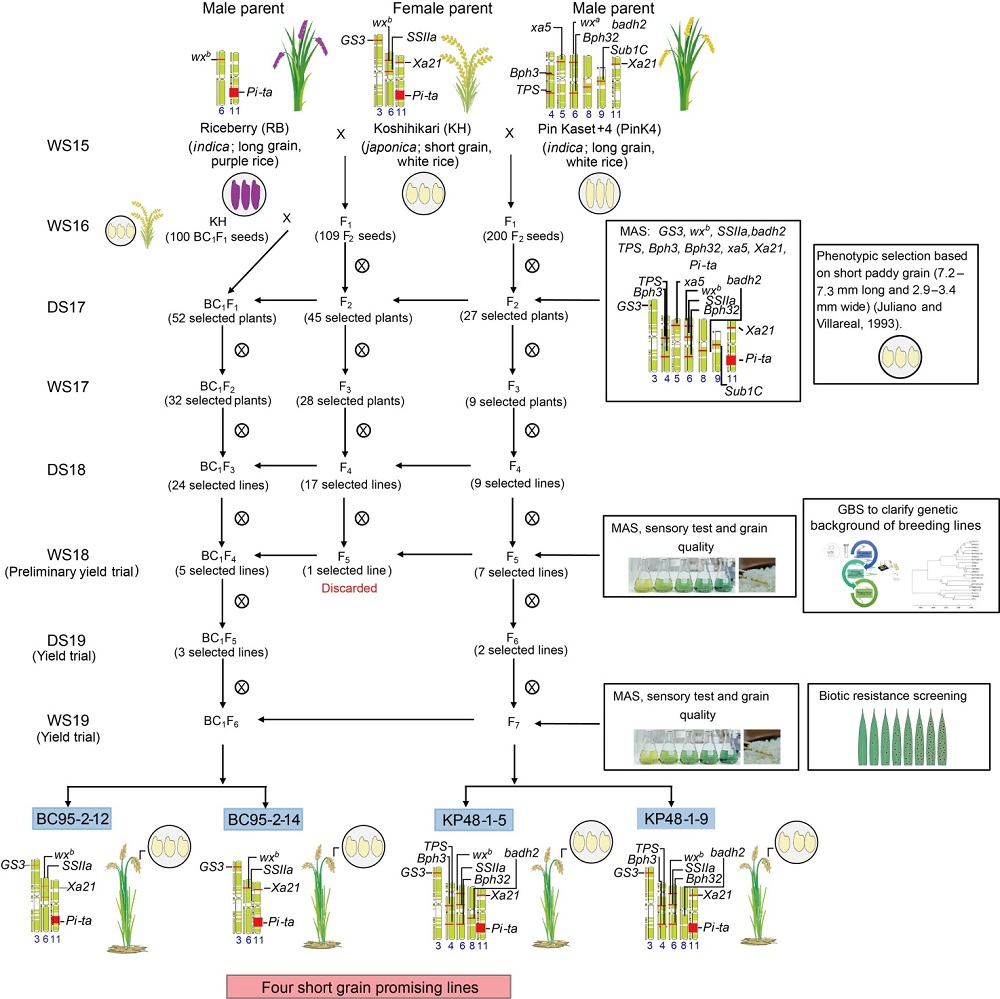
Fig. 1. Scheme of breeding programs for short grain rice derived from Koshihikari × Riceberry and Koshihikari × Pin Kaset+4 from WS15 to WS19 in Phan district, Chiang Rai Province, Thailand.WS, Wet season; DS, Dry season; MAS, Marker-assisted selection; GBS, Genotype by sequencing.
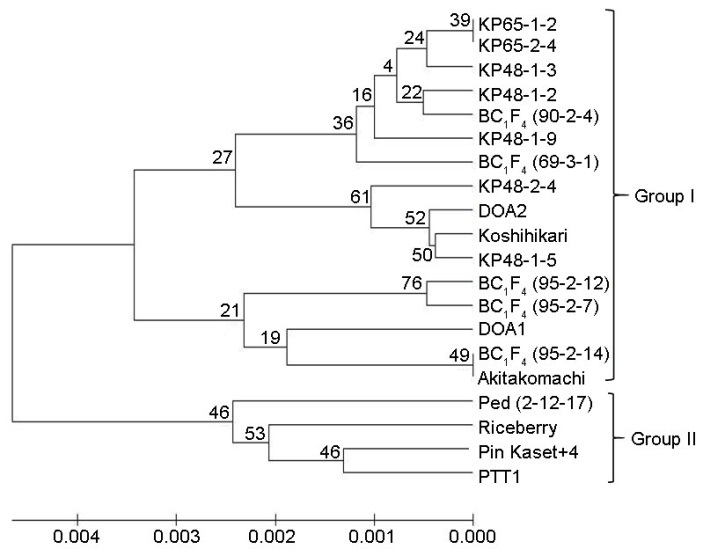
Fig. 3. Phylogenetic tree of breeding lines and control varieties based on genotyping by sequencing.The phylogenetic tree revealed two groups. Group I comprises the japonica type, while group II is made up of the indica type. The numbers at the node indicate the percentage obtained with 1000 bootstraps.
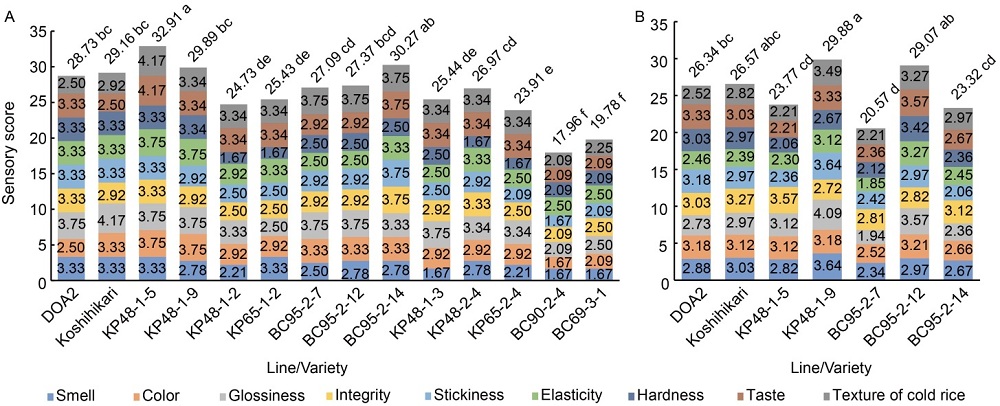
Fig. 4. Sensory test in BC1F5 derived from Koshihikari × Riceberry and F6 derived from Koshihikari × Pin Kaset+4 in wet season in 2018 (A) and candidate lines in BC1F6 and F7 in wet season in 2019 (B).Different lowercase letters follow the numbers above the column indicate significant differences among the lines at the 0.05 level using the LSD method.
| Line/Variety | GS3 | wxb | SSIIa | badh2 | Bph3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | |||||
| BC95-2-12 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| BC95-2-14 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| BC95-2-7 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| KP48-1-5 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
| KP48-1-9 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
| DOA1 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| DOA2 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| Koshihikari (KH) | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| Riceberry (RB) | -/- | -/- | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| Pin Kaset+4 (PinK4) | -/- | -/- | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | ||||
| Line/Variety | TPS | xa5 | Xa21 | Pi-ta | Sub1c | |||||||||
| WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | |||||
| BC95-2-12 | -/- | -/- | -/- | -/- | +/- | +/+ | +/+ | +/+ | -/- | -/- | ||||
| BC95-2-14 | -/- | -/- | -/- | -/- | +/- | +/+ | +/+ | +/+ | -/- | -/- | ||||
| BC95-2-7 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| KP48-1-5 | +/+ | +/+ | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| KP48-1-9 | +/+ | +/+ | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| DOA1 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| DOA2 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| Koshihikari (KH) | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| Riceberry (RB) | -/- | -/- | -/- | -/- | -/- | -/- | +/+ | +/+ | -/- | -/- | ||||
| Pin Kaset+4 (PinK4) | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
Table 3 SNP/InDel marker information on breeding lines identified in wet seasons of 2018 (WS18) and 2019 (WS19).
| Line/Variety | GS3 | wxb | SSIIa | badh2 | Bph3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | |||||
| BC95-2-12 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| BC95-2-14 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| BC95-2-7 | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| KP48-1-5 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
| KP48-1-9 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
| DOA1 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| DOA2 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| Koshihikari (KH) | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/- | -/- | -/- | -/- | ||||
| Riceberry (RB) | -/- | -/- | +/+ | +/+ | -/- | -/- | -/- | -/- | -/- | -/- | ||||
| Pin Kaset+4 (PinK4) | -/- | -/- | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | ||||
| Line/Variety | TPS | xa5 | Xa21 | Pi-ta | Sub1c | |||||||||
| WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | WS18 | WS19 | |||||
| BC95-2-12 | -/- | -/- | -/- | -/- | +/- | +/+ | +/+ | +/+ | -/- | -/- | ||||
| BC95-2-14 | -/- | -/- | -/- | -/- | +/- | +/+ | +/+ | +/+ | -/- | -/- | ||||
| BC95-2-7 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| KP48-1-5 | +/+ | +/+ | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| KP48-1-9 | +/+ | +/+ | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| DOA1 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| DOA2 | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| Koshihikari (KH) | -/- | -/- | -/- | -/- | +/+ | +/+ | +/+ | +/+ | -/- | -/- | ||||
| Riceberry (RB) | -/- | -/- | -/- | -/- | -/- | -/- | +/+ | +/+ | -/- | -/- | ||||
| Pin Kaset+4 (PinK4) | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||||
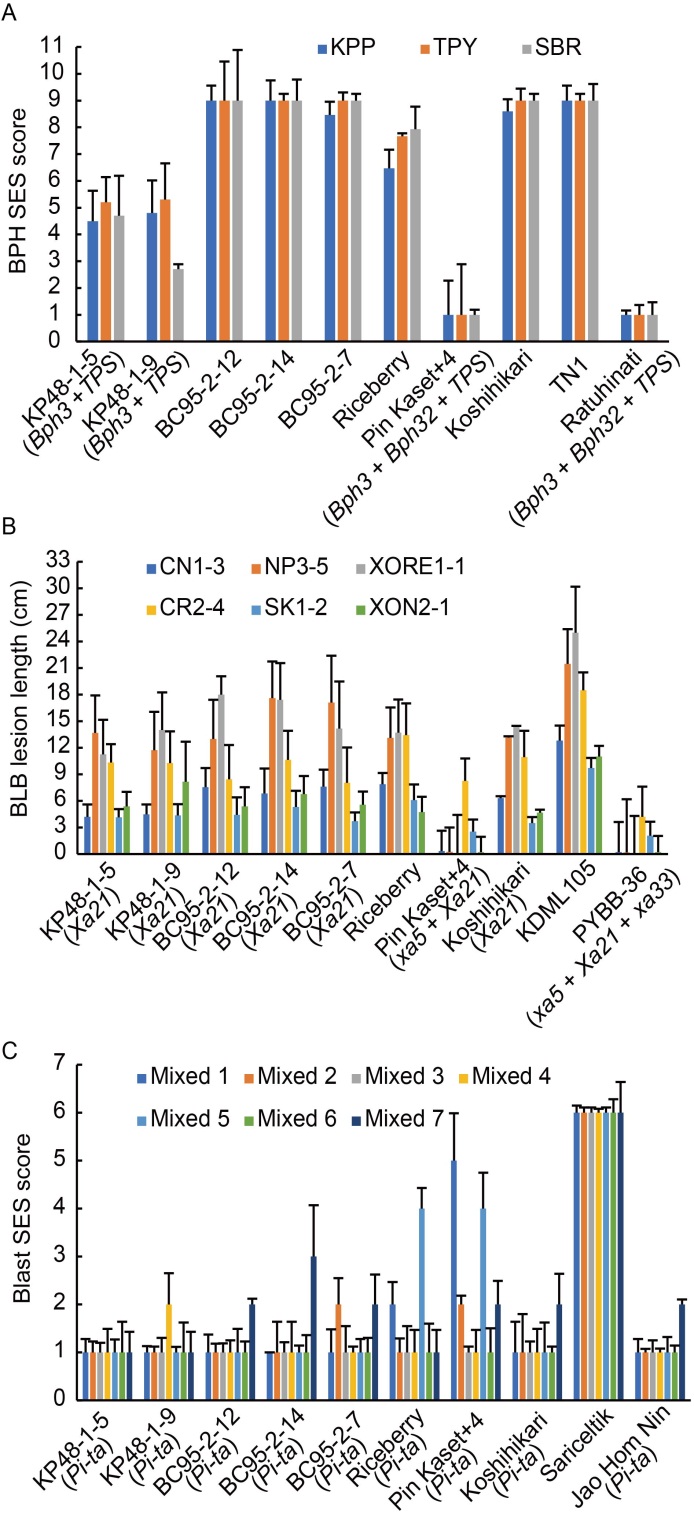
Fig. 5. Evaluation of candidate lines for brown planthopper (BPH), bacterial leaf blight (BLB) and blast resistance during wet season in 2019.A, Resistance of candidate lines, parents and control varieties against three biotypes of BPH. KPP, TPY and SBR refer to BPH populations of Kamphaeng Phet, Ta Phaya and Sing Buri, respectively. B, Resistance of candidate lines, parents and control varieties against six strains of BLB.C, Resistance of candidate lines, parents and control varieties against seven mixed strain groups of the blast. SES, Standard evaluation system. Data are Mean ± SD (n = 30).
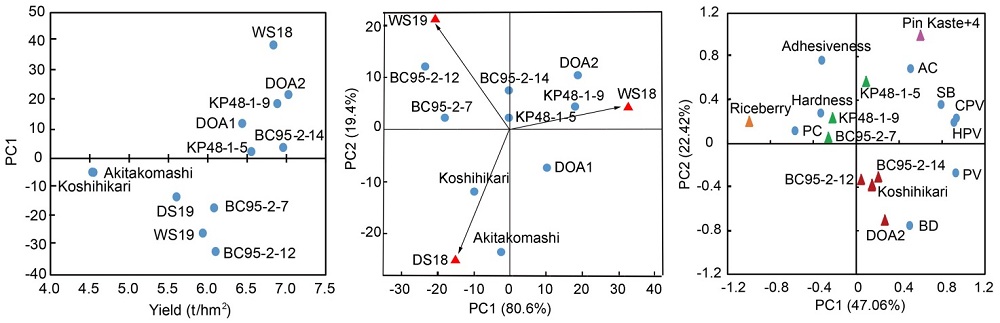
Fig. 6. Biplot graphs of grain yield and grain quality of five candidate lines and their control varieties.A, Biplot graph of the PC1 score versus the mean grain yield of five candidate lines and the control varieties in WS18, DS19 and WS19.B, Biplot graph of the PC1 score versus the PC2 score for the grain yields of five candidate lines and the control varieties in WS18, DS19 and WS19.C, Biplot graph of the PC1 score versus the PC2 score for the grain quality of five candidate lines and the control varieties. AC, Amylose content; SB, Setback; CPV, Cold paste viscosity; HPV, Hot paste viscosity; PV, Peak viscosity; BD, Breakdown; PC, Protein content.WS18, DS19 and WS19 refer to wet season in 2018, dry season in 2019 and wet season in 2019, respectively.
| Line/ Variety | Protein content (%) | Cooking time (min) | Viscosity (RVU) (Mean ± SD) | Cooked rice texture | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak viscosity | Hot paste viscosity | Breakdown | Cool paste viscosity | Setback | Hardness (N) | Stickiness (N∙s) | ||||
| BC95-2-12 | 6.88 b | 13.0 b | 216.96 ± 6.66 b | 120.21 ± 4.65 c | 96.75 ± 2.01 a | 191.88 ± 5.95 e | 71.67 ± 1.29 d | 32.06 bc | 19.30 bcd | |
| BC95-2-14 | 6.26 b | 22.5 a | 209.50 ± 1.88 b | 115.96 ± 1.47 c | 93.55 ± 3.36 ab | 202.21 ± 1.24 de | 86.25 ± 0.24 ab | 23.76 c | 20.99 bc | |
| BC95-2-7 | 5.43 b | 11.5 b | 137.08 ± 7.66 c | 89.79 ± 2.77 d | 47.29 ± 4.89 d | 163.83 ± 4.36 f | 74.04 ± 1.59 d | 45.50 ab | 21.91 bc | |
| KP48-1-5 | 8.51 a | 14.0 b | 204.55 ± 2.65 b | 159.75 ± 0.11 b | 44.79 ± 2.53 de | 250.71 ± 1.94 b | 90.96 ± 1.82 a | 47.39 a | 6.44 cd | |
| KP48-1-9 | 6.48 b | 10.5 b | 128.79 ± 2.53 c | 94.16 ± 1.29 d | 34.62 ± 1.24 ef | 176.17 ± 2.12 f | 82.00 ± 0.82 bc | 31.91 bc | 13.11 cd | |
| DOA2 | 6.87 b | 11.0 b | 232.00 ± 1.77 a | 150.30 ± 1.59 b | 81.71 ± 0.18 b | 227.21 ± 4.07 c | 76.92 ± 2.47 cd | 30.81 bc | 54.45 a | |
| Koshihikari | 6.07 b | 12.5 b | 213.92 ± 0.83 b | 152.04 ± 3.83 b | 61.88 ± 4.66 c | 224.55 ± 0.53 c | 72.50 ± 3.30 d | 31.74 bc | 32.80 b | |
| Pin Kaset+4 | 5.44 b | 13.0 b | 218.67 ± 0.71 ab | 191.38 ± 0.42 a | 27.29 ± 1.12 f | 282.08 ± 3.06 a | 90.71 ± 3.48 a | 26.19 c | 5.21 cd | |
| Riceberry | 8.81 a | 23.0 a | 59.34 ± 1.89 d | 51.55 ± 1.60 e | 7.79 ± 0.30 g | 102.08 ± 2.12 g | 54.05 ± 1.24 e | 33.43 bc | 2.53 d | |
Table 4 Physicochemical and cooking qualities of candidate lines compared with their parents and commercial varieties in wet season in 2019.
| Line/ Variety | Protein content (%) | Cooking time (min) | Viscosity (RVU) (Mean ± SD) | Cooked rice texture | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak viscosity | Hot paste viscosity | Breakdown | Cool paste viscosity | Setback | Hardness (N) | Stickiness (N∙s) | ||||
| BC95-2-12 | 6.88 b | 13.0 b | 216.96 ± 6.66 b | 120.21 ± 4.65 c | 96.75 ± 2.01 a | 191.88 ± 5.95 e | 71.67 ± 1.29 d | 32.06 bc | 19.30 bcd | |
| BC95-2-14 | 6.26 b | 22.5 a | 209.50 ± 1.88 b | 115.96 ± 1.47 c | 93.55 ± 3.36 ab | 202.21 ± 1.24 de | 86.25 ± 0.24 ab | 23.76 c | 20.99 bc | |
| BC95-2-7 | 5.43 b | 11.5 b | 137.08 ± 7.66 c | 89.79 ± 2.77 d | 47.29 ± 4.89 d | 163.83 ± 4.36 f | 74.04 ± 1.59 d | 45.50 ab | 21.91 bc | |
| KP48-1-5 | 8.51 a | 14.0 b | 204.55 ± 2.65 b | 159.75 ± 0.11 b | 44.79 ± 2.53 de | 250.71 ± 1.94 b | 90.96 ± 1.82 a | 47.39 a | 6.44 cd | |
| KP48-1-9 | 6.48 b | 10.5 b | 128.79 ± 2.53 c | 94.16 ± 1.29 d | 34.62 ± 1.24 ef | 176.17 ± 2.12 f | 82.00 ± 0.82 bc | 31.91 bc | 13.11 cd | |
| DOA2 | 6.87 b | 11.0 b | 232.00 ± 1.77 a | 150.30 ± 1.59 b | 81.71 ± 0.18 b | 227.21 ± 4.07 c | 76.92 ± 2.47 cd | 30.81 bc | 54.45 a | |
| Koshihikari | 6.07 b | 12.5 b | 213.92 ± 0.83 b | 152.04 ± 3.83 b | 61.88 ± 4.66 c | 224.55 ± 0.53 c | 72.50 ± 3.30 d | 31.74 bc | 32.80 b | |
| Pin Kaset+4 | 5.44 b | 13.0 b | 218.67 ± 0.71 ab | 191.38 ± 0.42 a | 27.29 ± 1.12 f | 282.08 ± 3.06 a | 90.71 ± 3.48 a | 26.19 c | 5.21 cd | |
| Riceberry | 8.81 a | 23.0 a | 59.34 ± 1.89 d | 51.55 ± 1.60 e | 7.79 ± 0.30 g | 102.08 ± 2.12 g | 54.05 ± 1.24 e | 33.43 bc | 2.53 d | |
| Variable | Peak viscosity | Hot paste viscosity | Breakdown | Cool paste viscosity | Setback | Amylose content | Protein content | Hardness | Adhesiveness |
|---|---|---|---|---|---|---|---|---|---|
| Hot paste viscosity | 0.847** | ||||||||
| Breakdown | 0.713** | 0.232 | |||||||
| Cool paste viscosity | 0.845** | 0.987** | 0.245 | ||||||
| Setback | 0.625* | 0.718** | 0.197 | 0.822** | |||||
| Amylose content | 0.228 | 0.511* | -0.256 | 0.536* | 0.522* | ||||
| Protein content | -0.408 | -0.319 | -0.327 | -0.351 | -0.350 | -0.393 | |||
| Hardness | -0.295 | -0.144 | -0.350 | -0.125 | -0.033 | -0.182 | 0.380 | ||
| Adhesiveness | -0.461 | -0.210 | -0.567 | -0.168 | 0.051 | 0.331 | 0.260 | 0.121 | |
| Cooking time | -0.353 | -0.438 | -0.069 | -0.442 | -0.322 | -0.185 | 0.409 | -0.299 | 0.382 |
Table 5 Correlation coefficients (r) of factors for all rice samples.
| Variable | Peak viscosity | Hot paste viscosity | Breakdown | Cool paste viscosity | Setback | Amylose content | Protein content | Hardness | Adhesiveness |
|---|---|---|---|---|---|---|---|---|---|
| Hot paste viscosity | 0.847** | ||||||||
| Breakdown | 0.713** | 0.232 | |||||||
| Cool paste viscosity | 0.845** | 0.987** | 0.245 | ||||||
| Setback | 0.625* | 0.718** | 0.197 | 0.822** | |||||
| Amylose content | 0.228 | 0.511* | -0.256 | 0.536* | 0.522* | ||||
| Protein content | -0.408 | -0.319 | -0.327 | -0.351 | -0.350 | -0.393 | |||
| Hardness | -0.295 | -0.144 | -0.350 | -0.125 | -0.033 | -0.182 | 0.380 | ||
| Adhesiveness | -0.461 | -0.210 | -0.567 | -0.168 | 0.051 | 0.331 | 0.260 | 0.121 | |
| Cooking time | -0.353 | -0.438 | -0.069 | -0.442 | -0.322 | -0.185 | 0.409 | -0.299 | 0.382 |

Fig. S2. Phenotypes of four promising lines and their parents in wet season in 2019.A, Plant type. B, Alkaline test. C, Cooked grains. D, Milled grains. E, Brown grains. F, Paddy grains of four promising lines and their parents.
| Target trait | Gene | Chr. | Marker name | SNP/InDel | Homozygous target SNP/InDel | LGC code a |
|---|---|---|---|---|---|---|
| Amylose content | wxb | 6 | wx_5UTR_G/T | G/T | T:T | 002-0052.1 |
| Gelatinization temperature | SSIIa | 6 | ALK_ex8_SNP_A/G | A/G | A:A | 002-0049.1 |
| Short grain | GS3 | GS3 | C/A | C:C | 002-0755.1 | |
| Aroma | badh2 | 8 | Aroma_2-3 | AAAAGATTATGGC/TATAT | TATAT:TATAT | 002-0829.1 |
| Blast resistance | Pi-ta | 11 | TBGI453598 | T/C | T:T | 002-0821.1 |
| Brown planthopper | Bph3 | 4 | OsLecRK3_QBPHR | A/G | G:G | 002-0263.1 |
| TPS | 4 | OsSTPS2_21bp_del | TTTATGCCTCTGGTGTGACCA/- | TTTATGCCTCTGGTGTGACCA:TTTATGCCTCTGGTGTGACCA | 002-0120.1 | |
| Bacterial leaf blight | xa5 | 5 | SNP_P98 | A/T | A:A | 002-0775.1 |
| Xa21 | 11 | SNP_P100 | G/A | G:G | 002-0998.1 | |
| Submerge | Sub1C | 9 | Sub1C_loci5 | T/C | T:T | 002-0995.1 |
Table S1 Gene-based/linked markers used for foreground selection of biotic resistance, abiotic tolerance and cooking quality for their validation in the breeding lines.
| Target trait | Gene | Chr. | Marker name | SNP/InDel | Homozygous target SNP/InDel | LGC code a |
|---|---|---|---|---|---|---|
| Amylose content | wxb | 6 | wx_5UTR_G/T | G/T | T:T | 002-0052.1 |
| Gelatinization temperature | SSIIa | 6 | ALK_ex8_SNP_A/G | A/G | A:A | 002-0049.1 |
| Short grain | GS3 | GS3 | C/A | C:C | 002-0755.1 | |
| Aroma | badh2 | 8 | Aroma_2-3 | AAAAGATTATGGC/TATAT | TATAT:TATAT | 002-0829.1 |
| Blast resistance | Pi-ta | 11 | TBGI453598 | T/C | T:T | 002-0821.1 |
| Brown planthopper | Bph3 | 4 | OsLecRK3_QBPHR | A/G | G:G | 002-0263.1 |
| TPS | 4 | OsSTPS2_21bp_del | TTTATGCCTCTGGTGTGACCA/- | TTTATGCCTCTGGTGTGACCA:TTTATGCCTCTGGTGTGACCA | 002-0120.1 | |
| Bacterial leaf blight | xa5 | 5 | SNP_P98 | A/T | A:A | 002-0775.1 |
| Xa21 | 11 | SNP_P100 | G/A | G:G | 002-0998.1 | |
| Submerge | Sub1C | 9 | Sub1C_loci5 | T/C | T:T | 002-0995.1 |
| Stage | Temperature | Duration | Number of cycles |
|---|---|---|---|
| 1 | 94 °C | 15 min | 1 |
| 2 | 94 °C | 20 s | 10 |
| 61 °C (decrease of 0.6 °C per cycle to achieve a final temperature of 55 °C) | 1 min | ||
| 3 | 94 °C | 20 s | 26 |
| 55 °C | 1 min |
Table S2 Thermal cycling conditions for polymerase chain reaction amplification used by HydrocyclerTM.
| Stage | Temperature | Duration | Number of cycles |
|---|---|---|---|
| 1 | 94 °C | 15 min | 1 |
| 2 | 94 °C | 20 s | 10 |
| 61 °C (decrease of 0.6 °C per cycle to achieve a final temperature of 55 °C) | 1 min | ||
| 3 | 94 °C | 20 s | 26 |
| 55 °C | 1 min |
| Mixed a | Isolate | Province of origin |
|---|---|---|
| 1 | THL211 | Chiang Mai, Thailand |
| THL137 | Chiang Mai, Thailand | |
| THL759 | Mae Hong Son, Thailand | |
| THL831 | Mae Hong Son, Thailand | |
| THL832 | Mae Hong Son, Thailand | |
| THL234 | Pathum Thani, Thailand | |
| 2 | THL710 | Mae Hong Son, Thailand |
| THL279 | Phrae, Thailand | |
| THL906 | Yala, Thailand | |
| THL881 | Chumphon, Thailand | |
| THL757 | Mae Hong Son, Thailand | |
| 3 | THL191 | Phitsanulok, Thailand |
| THL266 | Lampang, Thailand | |
| THL653 | Chiang Mai, Thailand | |
| THL658 | Chiang Rai, Thailand | |
| THL730 | Mae Hong Son, Thailand | |
| THL734 | Mae Hong Son, Thailand | |
| 4 | THL374 | Nakorn Ratchasima, Thailand |
| THL456 | Sakon Nakon, Thailand | |
| THL810 | Ubon Ratchathani, Thailand | |
| THL838 | Sri Saket, Thailand | |
| THL967 | Surin, Thailand | |
| THL985 | Nongkhai, Thailand | |
| 5 | THL144 | Chaing Mai, Thailand |
| THL284 | Nan, Thailand | |
| THL364 | Nakorn Ratchasima, Thailand | |
| THL690 | Lampang, Thailand | |
| THL1023 | Phayao, Thailand | |
| 6 | THL041 | Phitsanulok, Thailand |
| THL855 | Prachin Buri, Thailand | |
| THL949 | Suphan Buri, Thailand | |
| THL1003 | Bangkok, Thailand | |
| THL1009 | Sa Kaeo, Thailand | |
| 7 | TRG1 | Nongkhai, Thailand |
| TRG2 | Nongkhai, Thailand | |
| TRG17 | Lampang, Thailand | |
| Blast 4 | Lampang, Thailand | |
| TH196031 | Ubon Ratchathani, Thailand | |
| THL196036 | Ubon Ratchathani, Thailand |
Table S3 Seven mixed strain groups of blast diseases in Thailand as classified by AFLP.
| Mixed a | Isolate | Province of origin |
|---|---|---|
| 1 | THL211 | Chiang Mai, Thailand |
| THL137 | Chiang Mai, Thailand | |
| THL759 | Mae Hong Son, Thailand | |
| THL831 | Mae Hong Son, Thailand | |
| THL832 | Mae Hong Son, Thailand | |
| THL234 | Pathum Thani, Thailand | |
| 2 | THL710 | Mae Hong Son, Thailand |
| THL279 | Phrae, Thailand | |
| THL906 | Yala, Thailand | |
| THL881 | Chumphon, Thailand | |
| THL757 | Mae Hong Son, Thailand | |
| 3 | THL191 | Phitsanulok, Thailand |
| THL266 | Lampang, Thailand | |
| THL653 | Chiang Mai, Thailand | |
| THL658 | Chiang Rai, Thailand | |
| THL730 | Mae Hong Son, Thailand | |
| THL734 | Mae Hong Son, Thailand | |
| 4 | THL374 | Nakorn Ratchasima, Thailand |
| THL456 | Sakon Nakon, Thailand | |
| THL810 | Ubon Ratchathani, Thailand | |
| THL838 | Sri Saket, Thailand | |
| THL967 | Surin, Thailand | |
| THL985 | Nongkhai, Thailand | |
| 5 | THL144 | Chaing Mai, Thailand |
| THL284 | Nan, Thailand | |
| THL364 | Nakorn Ratchasima, Thailand | |
| THL690 | Lampang, Thailand | |
| THL1023 | Phayao, Thailand | |
| 6 | THL041 | Phitsanulok, Thailand |
| THL855 | Prachin Buri, Thailand | |
| THL949 | Suphan Buri, Thailand | |
| THL1003 | Bangkok, Thailand | |
| THL1009 | Sa Kaeo, Thailand | |
| 7 | TRG1 | Nongkhai, Thailand |
| TRG2 | Nongkhai, Thailand | |
| TRG17 | Lampang, Thailand | |
| Blast 4 | Lampang, Thailand | |
| TH196031 | Ubon Ratchathani, Thailand | |
| THL196036 | Ubon Ratchathani, Thailand |
| [1] | AACC. 2000. Approved Methods of the American Association of Cereal Chemists. 10th edn. St. Paul, MN, USA: America Association of Cereal Chemists, John Wiley & Sons Inc. |
| [2] | Allard R W. 1960. Principles of Plant Breeding. New York, USA: John Wiley & Sons Inc. |
| [3] | Chun A, Lee H J, Hamaker B R, Janaswamy S. 2015. Effects of ripening temperature on starch structure and gelatinization, pasting, and cooking properties in rice (Oryza sativa). J Agric Food Chem, 63: 3085-3093. |
| [4] | Das G, Rao G J. 2015. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front Plant Sci, 6: 698. |
| [5] | Gauch H G. 1988. Model selection and validation for yield trials with interaction. Biometrics, 44: 705-715. |
| [6] | Hamaker B R, Griffin V K. 1993. Effect of disulfide bond-containing protein on rice starch gelatinization and pasting. Cereal Chem, 70: 377-380. |
| [7] | Heinrichs E A, Medrano F G, Rapusas H R. 1985. Genetic Evaluation for Insect Resistance in Rice. Los Banos, the Philippines: International Rice Research Institute. |
| [8] | Hori K, Yamamoto T, Yano M. 2017. Genetic dissection of agronomically important traits in closely related temperatejaponica rice cultivars. Breeding Sci, 67: 427-434. |
| [9] | Hosoi N. 1979. Studies on meteorological fluctuation in the growth of rice plants: III. Relation between the heading response of rice varieties to temperature under natural daylength and the thermos- sensitivity, photosensitivity, basic vegetative growth under controlled environments. Jpn J Breeding, 29: 294-304. (in Japanese with English abstract) |
| [10] | International Rice Research Institute (IRRI). 2013. Standard Evaluation System (SES) for Rice. 5th edn. Los Banos, Manila, the Philippines: International Rice Research Institute: 46. |
| [11] | Ise K, Akama Y, Horisue N, Nakane A, Yokoo M, Ando I, Hata T, Sito M, Numaguchi K, Nemoto H. 2001. ‘Milky queen’ a new high-quality rice cultivar with low amylose content in endosperm. Bull Natl Inst Crop Sci, 2: 39-61. (in Japanese with English abstract) |
| [12] | Ishizaka S, Uehara Y, Fujita Y, Okuno K, Horiuchi H, Miura K, Nakagahra M, Yamada T, Uchiyamada H, Samoto S. 1989. Breeding process and characteristic of new released variety Kinuhikari. Hokuriku Crop Sci, 24: 25-27. (in Japanese) |
| [13] | Juliano B O. 1985. Criteria and test for rice grain quality. In: Rice Chemistry and Technology. Saint Paul, USA: American Association of Cereal Chemists (AACC): 443-513. |
| [14] | Juliano B O, Villareal C P. 1993. Grain Quality Evaluation of World Rice. Manila, the Philippines: International Rice Research Institute. |
| [15] | Kang K H. 2010. Made for the TROPICS. Rice Today, 9: 34-35. |
| [16] | Kobayashi A, Hori K, Yamamoto T, Yano M. 2018. Koshihikari: A premium short-grain rice cultivar: Its expansion and breeding in Japan. Rice, 11: 15. |
| [17] | Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods, 9: 357-359. |
| [18] | Lee J S, Torollo G, Ndayiragije A, Bizimana J B, Choi I R, Gulles A, Yeo U S, Jeong O Y, Venkatanagappa S, Kim B K. 2018. Genetic relationship of tropical region-bred temperatejaponica rice(Oryza sativa) plants and their grain yield variations in three different tropical environments. Plant Breeding, 137: 857-864. |
| [19] | LGC Group. 2016. SNPline genotyping automation. . (Accessed 5 January 2020) |
| [20] | Li H, Prakash S, Nicholson T M, Fitzgerald M A, Gilbert R G. 2016. The importance of amylose and amylopectin fine structure for textural properties of cooked rice grains. Food Chem, 196: 702-711. |
| [21] | Marchetti M A, Lai X, Bollich C N. 1987. Inheritance of resistance toPyricularia oryzae in rice cultivar grown in the United States. Phytopathology, 77: 799-804. |
| [22] | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. 2010. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res, 20: 1297-1303. |
| [23] | Miyamoto M. 2017. Influence of changing Thai society on the Japanese restaurant industry in Thailand. Manusya: J Hum, 20: 13-32. |
| [24] | Nakamura S, Okadome H, Yoza K, Haraguchi K, Okunishi T, Suzuki K, Sato H, Ohtsubo K. 2004. Differentiation and search for palatability-factors of world-wide rice grains by PCR method. J Jpn Soc Food Sci, 78: 764-779. (in Japanese with English abstract) |
| [25] | Nakwilai P, Cheabu S, Narumon P, Saensuk C, Arikita S, Malumpong C. 2020. Evaluation of japonica rice(Oryza sativa L.) varieties and their improvement in terms of stability, yield and cooking quality by pure-line selection in Thailand. Sci Asia, 46: 157-168. |
| [26] | Poosri S, Thilavech T, Pasukamonset P, Suparpprom C, Adisakwattana S. 2019. Studies on Riceberry rice (Oryza sativa L.) extract on the key steps related to carbohydrate and lipid digestion and absorption: A new source of natural bioactive substances. NFS J, 17: 17-23. |
| [27] | Ruengphayak S, Chaichumpoo E, Phromphan S, Kamolsukyunyong W, Sukhaket W, Phuvanartnarubal E, Korinsak S, Korinsak S, Vanavichit A. 2015. Pseudo-backcrossing design for rapidly pyramiding multiple traits into a preferential rice variety. Rice, 8: 7. |
| [28] | Rybka K, Miyamoto M, Ando I, Saito A, Kawasaki S. 1997. High resolution mapping of the indica-derived rice blast resistance genes: II. Pi-ta2 and Pi-ta and a consideration of their origin. Mol Plant Microbe Interact, 10: 517-524. |
| [29] | Seemanon K, Yamao M, Hosono K. 2015. Production of Japanese rice through contract farming system in Wiang Pa Pao district, Chiang Rai Province, Thailand. Am J Rural Dev, 3(2): 41-51. |
| [30] | Saleh M, Meullenet J F. 2015. Cooked rice texture and rice flour pasting properties; impacted by rice temperature during milling. J Food Sci Technol, 52: 1602-1609. |
| [31] | Tao K, Yu W, Prakash S, Gibert R G. 2020. Investigating cooked rice textural properties by instrumental measurements. Food Sci Hum Wellness, 9(2): 130-135. |
| [32] | Theerayout T. 2009. Identification of microsatellite markers (SSR) linked to a new bacterial blight resistance gene xa33(t) in rice cultivar ‘Ba7’. Maejo Int J Sci Technol, 3: 235-247. |
| [33] | Tong C, Chen Y L, Tang F F, Xu F F, Huang Y, Chen H, Bao J S. 2014. Genetic diversity of amylose content and RVA pasting parameters in 20 rice accessions grown in Hainan, China. Food Chem, 161: 239-245. |
| [34] | Wada T, Yasui H, Inoue T, Tsubone M, Ogata T, Doi K, Yoshimura A, Matsue Y, 2013. Validation of QTLs for eating quality of japonica rice ‘Koshihikari’ using backcross inbred lines. Plant Prod Sci, 16(2): 131-140. |
| [35] | Warinrak B. 2013. Japonica Rice Production Technology in Thailand. Rice Department, Thailand: CRC. (in Thai) |
| [36] | Win K M, Korinsak S, Jantaboon J, Siangliw M, Lanceras- Siangliw J, Sirithunya P, Vanavichit A, Pantuwan G, Jongdee B, Sishiwong N, Toojinda T. 2012. Breeding the Thai jasmine rice variety KDML105 for non-age-related broad-spectrum resistance to bacterial blight disease based on combined marker-assisted and phenotypic selection. Field Crops Res, 137: 186-194. |
| [37] | Wonglom P, Watcharachaiyakup J, Patarapuwadol S, Kositratana W. 2015. Assessment of diversity among pathotype of Xanthomonas oryzae pv. oryzae prevalent in Thailand. Agric Sci J, 46(2): 165-175. (in Thai with English abstract) |
| [38] | Xu Y J, Ying Y N, Ouyang S H, Duan X L, Sun H, Jiang S K, Sun S C, Bao J S. 2018. Factors affecting sensory quality of cooked japonica rice. Rice Sci, 25(6): 330-339. |
| [39] | Yang Z, Sun X, Wang S, Zhang Q. 2003. Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor Appl Genet, 106: 1467-1472. |
| [40] | Yoshida S. 1983. Rice. In: Smith W H, Banta S J. Potential Productivity of Field Crops under Different Environments. Los Baños, the Philippines: International Rice Research Institute: 103-127. |
| [41] | Yugander A, Sundaram R M, Ladhalakshmi D, Hajira S K, Prakasam V, Prasad M S, Madhav M S, Babu V R, Laha G S. 2017. Virulence profiling of Xanthomonas oryzae pv. oryzae isolates, causing bacterial blight of rice in India. Eur J Plant Pathol, 149: 171-191. |
| [42] | Zhao C J, Xie J Q, Li L, Cao C J. 2017. Comparative transcriptomic analysis in the paddy rice under storage and identification of differentially regulated genes in response to high temperature and humidity. J Agric Food Chem, 65: 8145-8153. |
| [1] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [2] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [3] | Liu Yantong, Li Ting, Jiang Zhishu, Zeng Chuihai, He Rong, Qiu Jiao, Lin Xiaoli, Peng Limei, Song Yongping, Zhou Dahu, Cai Yicong, Zhu Changlan, Fu Junru, He Haohua, Xu Jie. Characterization of a Novel Weak Allele of RGA1/D1 and Its Potential Application in Rice Breeding [J]. Rice Science, 2022, 29(6): 522-534. |
| [4] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [5] | Luo Haowen, He Longxin, Du Bin, Pan Shenggang, Mo Zhaowen, Yang Shuying, Zou Yingbin, Tang Xiangru. Epoxiconazole Improved Photosynthesis, Yield Formation, Grain Quality and 2-Acetyl-1-Pyrroline Biosynthesis of Fragrant Rice [J]. Rice Science, 2022, 29(2): 189-196. |
| [6] | R. Abdul Fiyaz, D. Shivani, K. Chaithanya, K. Mounika, M. Chiranjeevi, G. S. Laha, B. C. Viraktamath, L. V. Subba Rao, R. M. Sundaram. Genetic Improvement of Rice for Bacterial Blight Resistance: Present Status and Future Prospects [J]. Rice Science, 2022, 29(2): 118-132. |
| [7] | Muduli Lakesh, Kumar Pradhan Sukanta, Mishra Abinash, Nath Bastia Debendra, Chandra Samal Kailash, Kumar Agrawal Pawan, Dash Manasi. Understanding Brown Planthopper Resistance in Rice: Genetics, Biochemical and Molecular Breeding Approaches [J]. Rice Science, 2021, 28(6): 532-546. |
| [8] | Panigrahy Madhusmita, Das Subhashree, Poli Yugandhar, Kumar Sahoo Pratap, Kumari Khushbu, C. S. Panigrahi Kishore. Carbon Nanoparticle Exerts Positive Growth Effects with Increase in Productivity by Down-Regulating Phytochrome B and Enhancing Internal Temperature in Rice [J]. Rice Science, 2021, 28(3): 289-300. |
| [9] | Jan Mehmood, Shah Gulmeena, Yuqing Huang, Xuejiao Liu, Peng Zheng, Hao Du, Hao Chen, Jumin Tu. Development of Heat Tolerant Two-Line Hybrid Rice Restorer Line Carrying Dominant Locus of OsHTAS [J]. Rice Science, 2021, 28(1): 99-108. |
| [10] | Yuyu Chen, Aike Zhu, Pao Xue, Xiaoxia Wen, Yongrun Cao, Beifang Wang, Yue Zhang, Liaqat Shah, Shihua Cheng, Liyong Cao, Yingxin Zhang. Effects of GS3 and GL3.1 for Grain Size Editing by CRISPR/Cas9 in Rice [J]. Rice Science, 2020, 27(5): 405-413. |
| [11] | B. ANGELES-SHIM Rosalyn, P. REYES Vincent, M. del VALLE Marilyn, S. LAPIS Ruby, SHIM Junghyun, SUNOHARA Hidehiko, K. JENA Kshirod, ASHIKARI Motoyuki, DOI Kazuyuki. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast [J]. Rice Science, 2020, 27(2): 113-123. |
| [12] | Chuan Tong, Lei Liu, L. E. Waters Daniel, Jin-song Bao. Association Mapping and Marker Development of Genes for Starch Lysophospholipid Synthesis in Rice [J]. Rice Science, 2016, 23(6): 287-296. |
| [13] | Ur Rehman Hafeez, Kamran Muhammad, Maqsood Ahmed Basra Shahzad, Afzal Irfan, Farooq Muhammad. Influence of Seed Priming on Performance and Water Productivity of Direct Seeded Rice in Alternating Wetting and Drying [J]. Rice Science, 2015, 22(4): 189-196. |
| [14] | Swar Oo Kyaw, Kongjaimun Alisa, Khanthong Srisawat, Yi Myint, Tin Myint Tin, Korinsak Siriporn, Lanceras Siangliw Jonaliza, Myo Myint Khin, Vanavichit Apichart, Malumpong Chanate, Toojinda Theerayut. Characterization of Myanmar Paw San Hmwe Accessions Using Functional Genetic Markers [J]. Rice Science, 2015, 22(2): 53-64. |
| [15] | LIU Qi-hua, WU Xiu, CHEN Bo-cong, MA Jia-qing, GAO Jie. Effects of Low Light on Agronomic and Physiological Characteristics of Rice Including Grain Yield and Quality [J]. RICE SCIENCE, 2014, 21(5): 243-251. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||