
Rice Science ›› 2024, Vol. 31 ›› Issue (2): 190-202.DOI: 10.1016/j.rsci.2023.12.002
• Research Papers • Previous Articles Next Articles
Wu Lijuan1,#, Han Cong1,#, Wang Huimei1, He Yuchang1, Lin Hai1, Wang Lei1, Chen Chen2( ), E Zhiguo1(
), E Zhiguo1( )
)
Received:2023-07-29
Accepted:2023-11-05
Online:2024-03-28
Published:2024-04-11
Contact:
E Zhiguo (About author:First author contact:#These authors contributed equally to this work
Wu Lijuan, Han Cong, Wang Huimei, He Yuchang, Lin Hai, Wang Lei, Chen Chen, E Zhiguo. OsbZIP53 Negatively Regulates Immunity Response by Involving in Reactive Oxygen Species and Salicylic Acid Metabolism in Rice[J]. Rice Science, 2024, 31(2): 190-202.
Add to citation manager EndNote|Ris|BibTeX
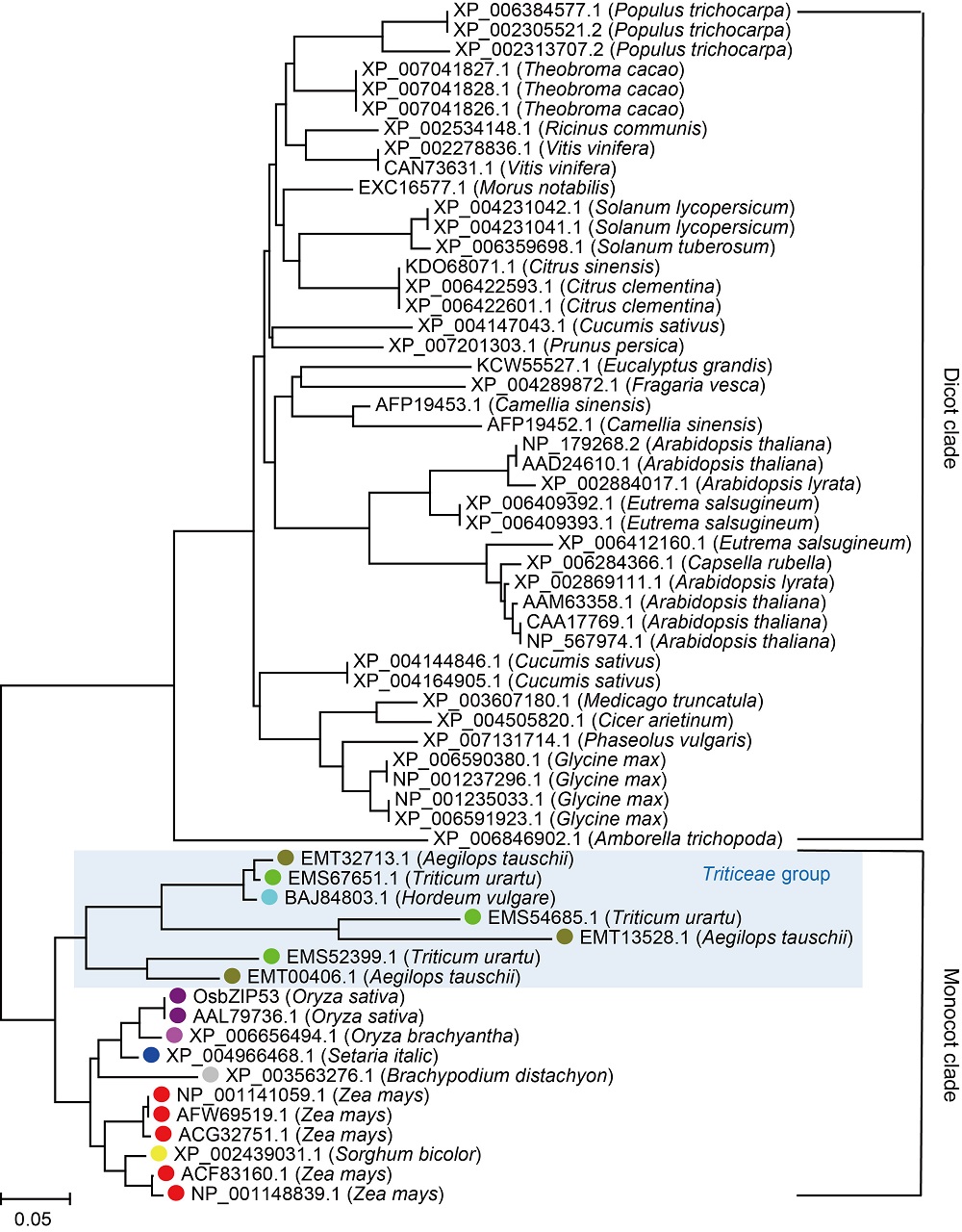
Fig. 1. Phylogenetic tree of OsbZIP53 and its homologs in plants. Homologs are indicated by NCBI accession numbers. Monocot homologs are marked with dots with distinctive color.
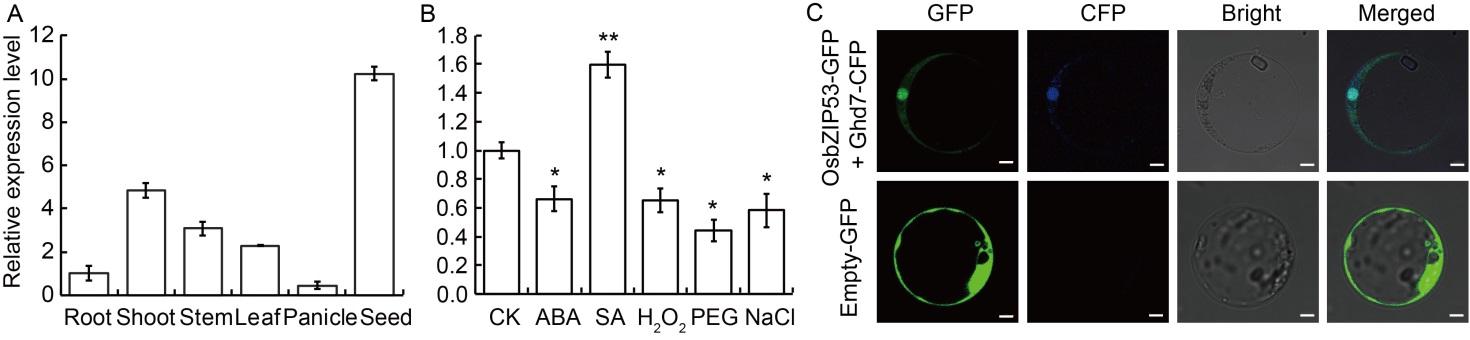
Fig. 2. Expression pattern and subcellular localization of OsbZIP53. A, Expression of OsbZIP53 in different rice tissues detected by qRT-PCR. Root and shoot were at 14 d after germination, stem and leaf were at 60 d after germination, panicle was 5‒8 cm in length, and seed was at 12 d after fertilization. The ACTIN gene was used as an internal standard, and relative expression levels were quantified using the 2-ΔΔCT threshold cycle method. Error bars indicated standard deviation. Three biological replicates were conducted for each sample. B, Expression of OsbZIP53 induced by different hormone and stress treatments. Seven-day-old rice seedlings were given the treatments (stress or hormones) for 3 h. CK, Control; ABA, 50 μmol/L abscisic acid; SA, 2 mmol/L salicylic acid; H2O2, 100 mmol/L H2O2; PEG, 20% polyethylene glycol 6000; NaCl, 100 mmol/L NaCl. Data were means and standard deviations of triplicates. The stars above the bars indicated the significant differences under treatments with respect to control in the t-test (*, P < 0.05; **, P ≤ 0.01). C, Subcellular localization of OsbZIP53 transiently expressed in rice protoplasts. Ghd7-CFP was used as a nuclear localization marker. GFP, Green fluorescent protein; CFP, Cyan fluorescent protein. Scale bars, 5 μm.
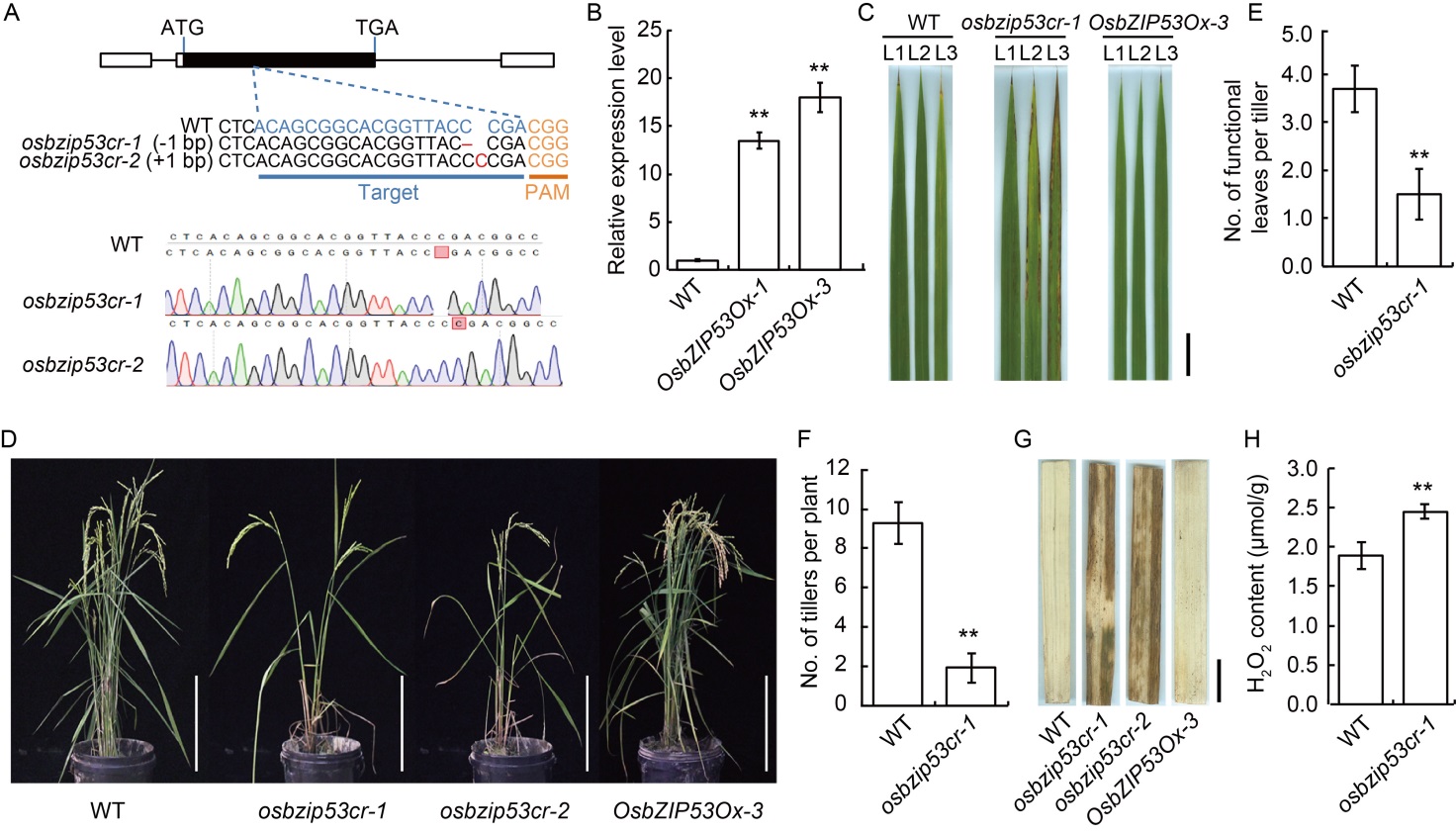
Fig. 3. Phenotypes of CRISPR/Cas9-induced OsbZIP53 mutants and over-expression transgenic plants. A, Schematic organization of the coding region of OsbZIP53, the sgRNA target site was underlined, the protospacer adjacent motif (PAM) was shown in orange, and the insertion and deletion mutations were indicated in red. The bottom was the sequencing results correspond to the mutation type. B, Relative expression of OsbZIP53 in wild type (WT) and over-expression transgenic lines (OsbZIP53Ox-1 and OsbZIP53Ox-3). The leaves from 8-week-old WT and OsbZIP53 mutant lines were used for qRT-PCR. The ACTIN gene was used as an internal standard, and relative expression levels were quantified using the 2-ΔΔCT threshold cycle method. Three biological replicates were employed for each sample.C, Morphology of different age leaves in WT and transgenic lines (osbzip53cr-1 and OsbZIP53Ox-3). L1, L2 and L3 indicated the upmost leaf, the second upmost leaf and the third upmost leaf, respectively. Scale bar, 2 cm.D, Whole plant morphology in WT and transgenic lines. Scale bars, 10 cm.E, Number of functional leaves on each tiller of osbzip53cr-1 and WT. F, Number of tillers per plant in osbzip53cr-1 and WT. G, 3,3ʹ-Diaminobenzidine (DAB) staining to show H2O2 accumulation in WT and transgenic plants. Scale bar, 1 cm.H, H2O2 content in the leaves of osbzip53cr-1 and WT. Data in B, E, F, and H are Mean ± SD (n = 3). ** (P ≤ 0.01) are detected by the t-test between the transgenic plants and WT.
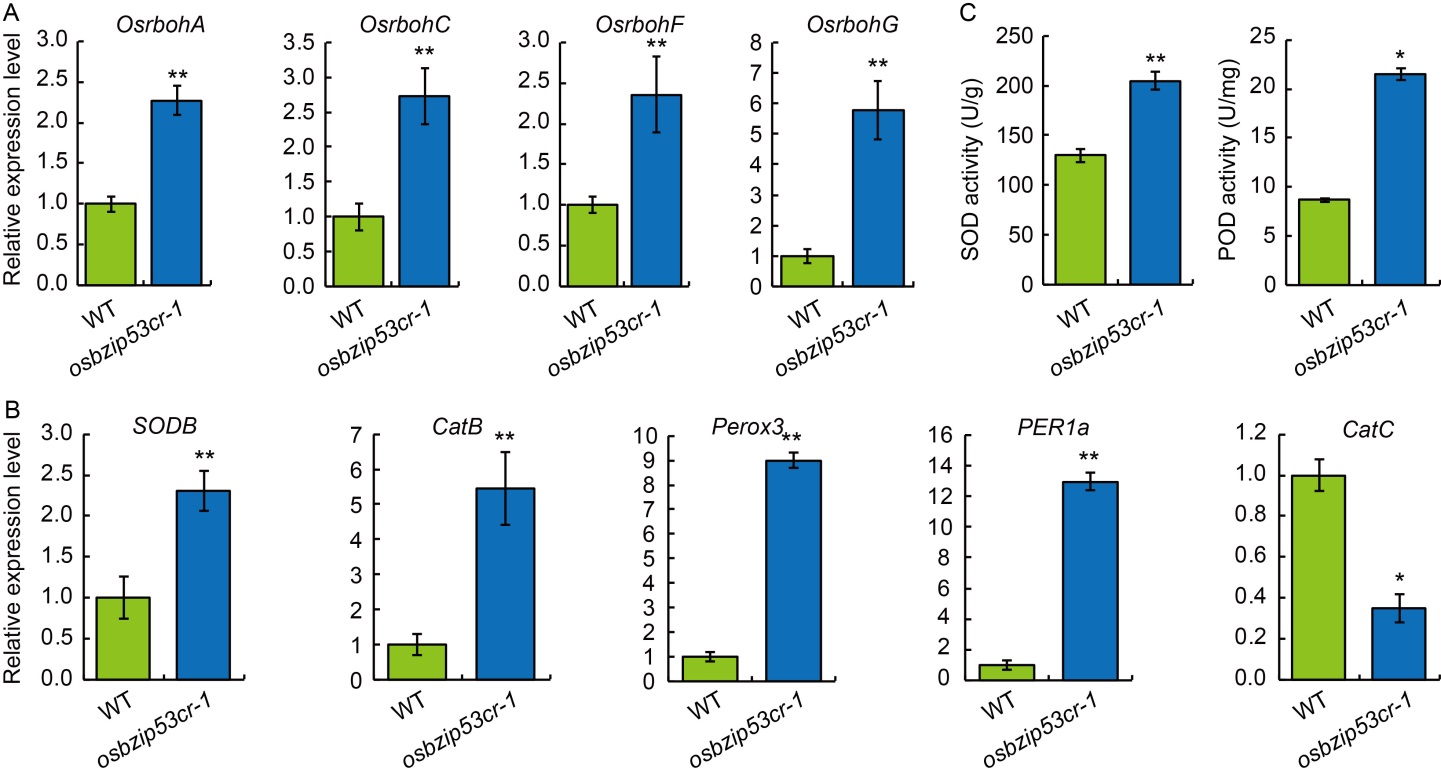
Fig. 4. Analysis of reactive oxygen species (ROS)-associated gene expression and enzyme activity. A, Relative expression levels of respiratory burst oxidase homolog revealed by qRT-PCR osbzip53cr mutant and wild type (WT). B, Relative expression levels of ROS-associated genes in osbzip53cr mutant and WT. C, Determination of superoxide dismutase (SOD) and peroxidase (POD) activities in osbzip53cr mutant and WT. The leaves from 6-week-old WT and osbzip53cr mutant lines were used for qRT-PCR. The ACTIN gene was used as an internal standard, and relative expression levels were quantified using the 2-ΔΔCT threshold cycle method. Data are Mean ± SD (n = 3). * (P < 0.05) and ** (P ≤ 0.01) indicate significant differences between osbzip53cr and WT in the t-test.
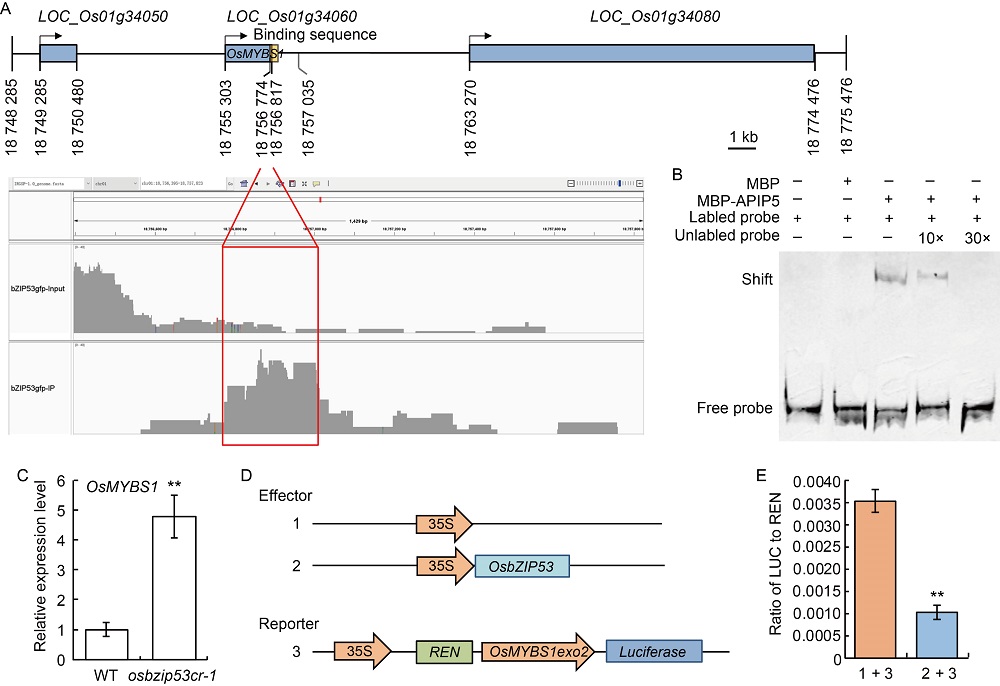
Fig. 5. OsMYBS1 may act as putative direct downstream target gene of OsbZIP53. A, Genome browser tracks showing OsbZIP53 binding signal around the OsMYBS1 locus by chromatin immunoprecipitation (ChIP)-seq. Blue boxes represent exons, yellow box represents binding sequence, and arrows represent transcription directions. B, Binding of maltose-binding protein (MBP)-OsbZIP53 to the probe around the OsMBYS1 locus in an electrophoretic mobility shift assay. MBP alone served as a control. The experiment was repeated three times.C, Relative expression levels of OsMBYS1 in osbzip53cr and wild type (WT). The leaves from 6-week-old WT and OsbZIP53 mutant lines were used for qRT-PCR. The ACTIN gene was used as an internal standard, and relative expression levels were quantified using the 2-ΔΔCT threshold cycle method.D, Diagram of constructs used in transcriptional activity assay. REN, Renilla. OsMYBS1exo2 indicates the sequence located on rice chromosome 1 at 18 756 774‒18 757 035 bp, which overlaps with the end of the second exon of OsMYBS1. E, Relative LUC (luciferase) activity represents the transcriptional activity. The activities of firefly LUC (LUC) and Renilla LUC (REN, as a control) were assayed, and the ratio of LUC/REN was calculated to represent the relative LUC activity. ‘1 + 3’ and ‘2 +3’ represent the combinations of effector 1 and reporter 3 and effector 2 and reporter 3 in D, respectively. Data in C and E are Mean ± SD (n = 3). ** indicates highly significant differences (P ≤ 0.01) detected by the t-test.
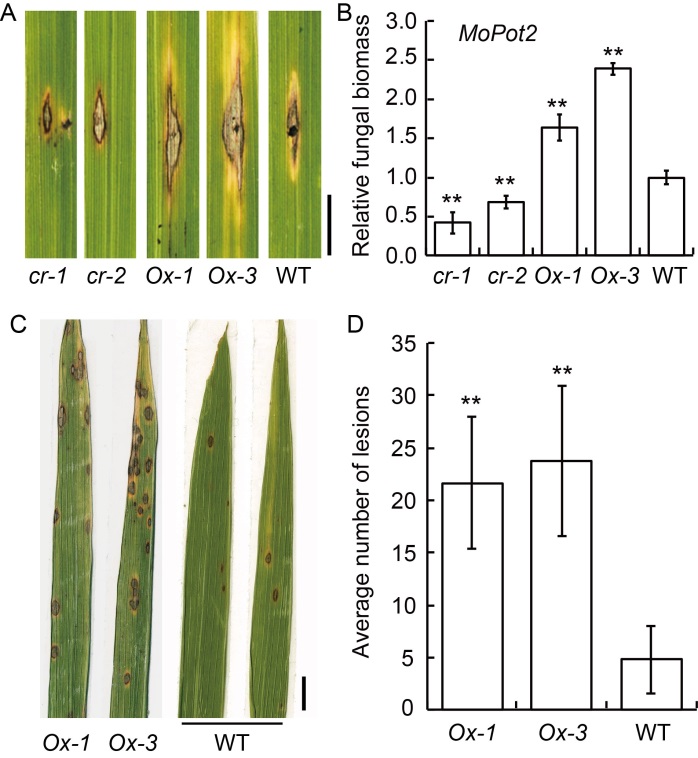
Fig. 6. Repression of OsbZIP53 increases resistance to Magnaporthe oryzae. A, Lesions in the leaves of osbzip53cr (cr-1 and cr-2), OsbZIP53Ox (Ox-1 and Ox-3), and wild type (WT) at 7 d after post-inoculation. Scale bar, 0.5 cm. B, Relative fungal biomass of osbzip53cr, OsbZIP53Ox and WT in A. The relative fungal biomass was estimated by DNA-based quantitative PCR using the CT value of M. oryzae transposable element MoPot2 relative to that of the rice Ubiquitin gene. C, Lesions in the leaves of OsbZIP53Ox and WT under natural disease state in the field. Scale bar, 1 cm. D, Average number of lesions formed in OsbZIP53Ox and WT under natural disease state in the field. Data are Mean ± SD (n = 3). ** indicates highly significant differences at P ≤ 0.01 detected by the t-test.
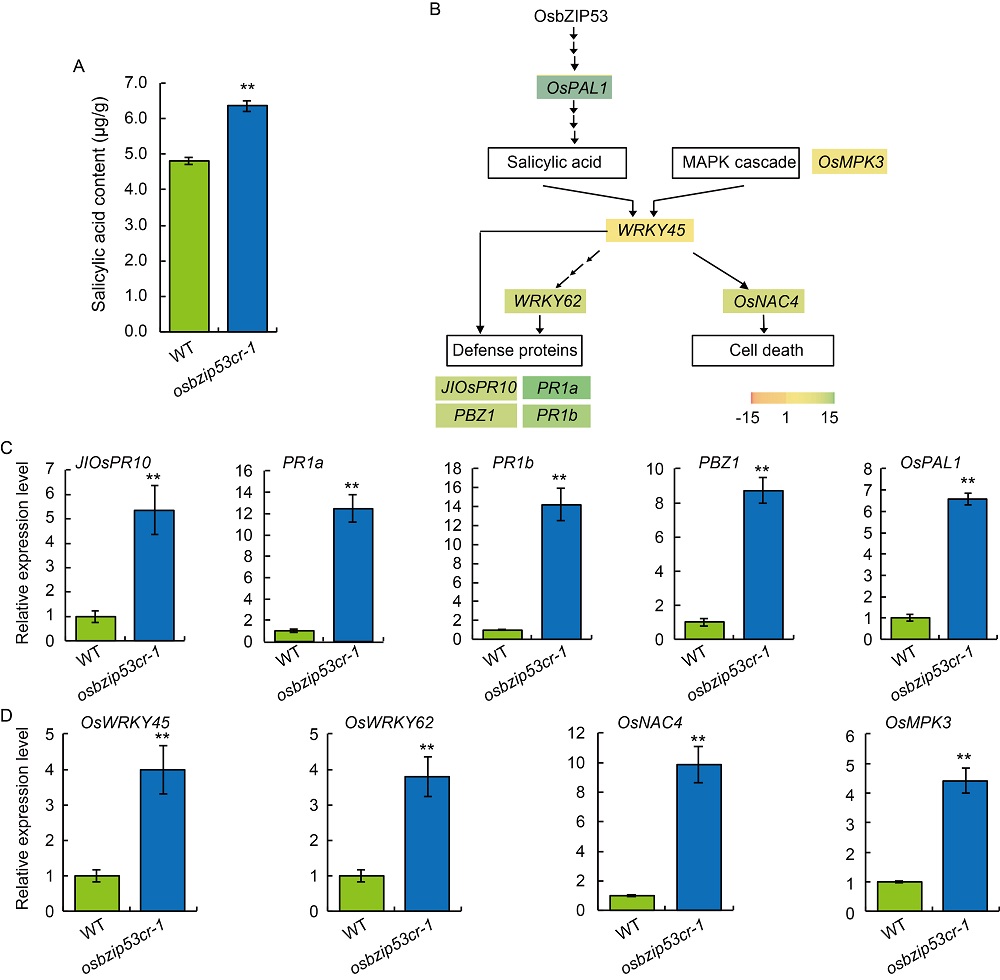
Fig. 7. Defense responses are activated in ObZIP53 mutant plants. A, Salicylic acid content in leaves of osbzip53cr-1 and wild type (WT). B, Schematic map of genes involved in processes related to defense response. The color scale of genes indicates the relative transcriptional level |log2 (fold change)| of the genes in osbzip53cr-1 in comparison to the WT plants based on RNA-seq results. MAPK, Mitogen-activated protein kinase.C and D, Relative expression levels of defense response-related genes (C) and OsWRKY45 pathway-related genes (D) revealed by qRT-PCR. Leaves from 6-week-old WT and OsbZIP53 mutant lines were used. The ACTIN gene was used as an internal standard, and relative expression levels were quantified using the 2-ΔΔCT threshold cycle method. Data are Mean ± SD (n = 3). ** indicates highly significant differences at P ≤ 0.01 detected by the t-test.
| [1] | Akamatsu A, Wong H L, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, Shimamoto K. 2013. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe, 13(4): 465-476. |
| [2] | Amoutzias G D, Veron A S, Weiner J III, Robinson-Rechavi M, Bornberg-Bauer E, Oliver S G, Robertson D L. 2007. One billion years of bZIP transcription factor evolution: Conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol, 24(3): 827-835. |
| [3] | Assunção A G L, Herrero E, Lin Y F, Huettel B, Talukdar S, Smaczniak C, Immink R G H, van Eldik M, Fiers M, Schat H, Aarts M G M. 2010. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA, 107(22): 10296-10301. |
| [4] | Chern M, Bai W, Ruan D L, Oh T, Chen X W, Ronald P C. 2014. Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins. BMC Genom, 15: 461. |
| [5] | Dröge-Laser W, Snoek B L, Snel B, Weiste C. 2018. The Arabidopsis bZIP transcription factor family: An update. Curr Opin Plant Biol, 45: 36-49. |
| [6] | Fang H, Shen S Q, Wang D, Zhang F, Zhang C Y, Wang Z X, Zhou Q Q, Wang R Y, Tao H, He F, Yang C K, Peng M, Jing X Y, Hao Z Y, Liu X L, Luo J, Wang G L, Ning Y S. 2021. A monocot-specific hydroxycinnamoylputrescine gene cluster contributes to immunity and cell death in rice. Sci Bull, 66(23): 2381-2393. |
| [7] | Fang H, Zhang F, Zhang C Y, Wang D, Shen S Q, He F, Tao H, Wang R Y, Wang M, Wang D B, Liu X L, Luo J, Wang G L, Ning Y S. 2022. Function of hydroxycinnamoyl transferases for the biosynthesis of phenolamides in rice resistance to Magnaporthe oryzae. J Genet Genomics, 49(8): 776-786. |
| [8] | Fitzgerald H A, Canlas P E, Chern M S, Ronald P C. 2005. Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae Plant J, 43(3): 335-347. |
| [9] | Han C, He Y C, Wu L J, Jia L L, Wang L, E Z G. 2023. Research progress on the function of basic leucine zipper (bZIP) protein family in rice. Chin J Rice Sci, 37(4): 436-448. (in Chinese with English abstract) |
| [10] | He Y, Shi Y F, Zhang X B, Xu X, Wang H M, Li L J, Zhang Z H, Shang H H, Wang Z H, Wu J L. 2020. The OsABCI7 transporter interacts with OsHCF222 to stabilize the thylakoid membrane in rice. Plant Physiol, 184(1): 283-299. |
| [11] | Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends Plant Sci, 7(3): 106-111. |
| [12] | Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong H L, Kawasaki T, Shimamoto K. 2010. Activation of a rac GTPase by the NLR family disease resistance protein pit plays a critical role in rice innate immunity. Cell Host Microbe, 7(5): 362-375. |
| [13] | Khanam S, Bauters L, Singh R R, Verbeek R, Haeck A, Sultan S M D, Demeestere K, Kyndt T, Gheysen G. 2018. Mechanisms of resistance in the rice cultivar Manikpukha to the rice stem nematode Ditylenchus angustus. Mol Plant Pathol, 19(6): 1391-1402. |
| [14] | Kim J S, Chae S, Jun K M, Pahk Y M, Lee T H, Chung P J, Kim Y K, Nahm B H. 2017. Genome-wide identification of grain filling genes regulated by the OsSMF1 transcription factor in rice. Rice, 10(1): 16. |
| [15] | Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 33(7): 1870-1874. |
| [16] | Li M M, Yao T, Lin W R, Hinckley W E, Galli M, Muchero W, Gallavotti A, Chen J G, Huang S S C. 2023. Double DAP-seq uncovered synergistic DNA binding of interacting bZIP transcription factors. Nat Commun, 14: 2600. |
| [17] | Li W T, Zhu Z W, Chern M, Yin J J, Yang C, Ran L, Cheng M P, He M, Wang K, Wang J, Zhou X G, Zhu X B, Chen Z X, Wang J C, Zhao W, Ma B T, Qin P, Chen W L, Wang Y P, Liu J L, Wang W M, Wu X J, Li P, Wang J R, Zhu L H, Li S G, Chen X W. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell, 170(1): 114-126. |
| [18] | Li Z Y, Wei X J, Tong X H, Zhao J, Liu X X, Wang H M, Tang L Q, Shu Y Z, Li G H, Wang Y F, Ying J Z, Jiao G A, Hu H H, Hu P S, Zhang J. 2022. The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol Plant, 15(4): 706-722. |
| [19] | Liu C T, Ou S J, Mao B G, Tang J Y, Wang W, Wang H R, Cao S Y, Schläppi M R, Zhao B R, Xiao G Y, Wang X P, Chu C C. 2018. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun, 9(1): 3302. |
| [20] | Nijhawan A, Jain M, Tyagi A K, Khurana J P. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol, 146(2): 333-350. |
| [21] | Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. 2009. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J Biol Chem, 284(39): 26510-26518. |
| [22] | Ruan B P, Hua Z H, Zhao J, Zhang B, Ren D Y, Liu C L, Yang S L, Zhang A P, Jiang H Z, Yu H P, Hu J, Zhu L, Chen G, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. 2019. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol J, 17(7): 1344-1356. |
| [23] | Seo Y S, Chern M, Bartley L E, Han M H, Jung K H, Lee I, Walia H, Richter T, Xu X, Cao P J, Bai W, Ramanan R, Amonpant F, Arul L, Canlas P E, Ruan R, Park C J, Chen X W, Hwang S, Jeon J S, Ronald P C. 2011. Towards establishment of a rice stress response interactome. PLoS Genet, 7(4): e1002020. |
| [24] | Song S, Wang G F, Wu H, Fan X W, Liang L W, Zhao H, Li S L, Hu Y, Liu H Y, Ayaad M, Xing Y Z. 2020. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J, 103(2): 532-546. |
| [25] | Sugio A, Yang B, Zhu T, White F F. 2007. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci USA, 104(25): 10720-10725. |
| [26] | Thordal-Christensen H, Zhang Z G, Wei Y D, Collinge D B. 1997. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J, 11(6): 1187-1194. |
| [27] | Thuan N T N, Bigirimana J, Roumen E, Van Der Straeten D, Höfte M. 2006. Molecular and pathotype analysis of the rice blast fungus in North Vietnam. Eur J Plant Pathol, 114(4): 381-396. |
| [28] | Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang C J, Goto S, Takahashi A, Hirochika H, Takatsuji H. 2013. MAP kinases phosphorylate rice WRKY45. Plant Signal Behav, 8(6): e24510. |
| [29] | Wamstad J A, Alexander J M, Truty R M, Shrikumar A, Li F G, Eilertson K E, Ding H M, Wylie J N, Pico A R, Capra J A, Erwin G, Kattman S J, Keller G M, Srivastava D, Levine S S, Pollard K S, Holloway A K, Boyer L A, Bruneau B G. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell, 151(1): 206-220. |
| [30] | Wang J Y, Wang R Y, Fang H, Zhang C Y, Zhang F, Hao Z Y, You X M, Shi X T, Park C H, Hua K Y, He F, Bellizzi M, Xuan Vo K T, Jeon J S, Ning Y S, Wang G L. 2021. Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol Plant, 14(2): 253-266. |
| [31] | Wang R Y, Ning Y S, Shi X T, He F, Zhang C Y, Fan J B, Jiang N, Zhang Y, Zhang T, Hu Y J, Bellizzi M, Wang G L. 2016. Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr Biol, 26(18): 2399-2411. |
| [32] | Wei K F, Chen J, Wang Y M, Chen Y H, Chen S X, Lin Y N, Pan S, Zhong X J, Xie D X. 2012. Genome-wide analysis of bZIP- encoding genes in maize. DNA Res, 19(6): 463-476. |
| [33] | Wong H L, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, Shimamoto K. 2007. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell, 19(12): 4022-4034. |
| [34] | Xue W Y, Xing Y Z, Weng X Y, Zhao Y, Tang W J, Wang L, Zhou H J, Yu S B, Xu C G, Li X H, Zhang Q F. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet, 40(6): 761-767. |
| [35] | Yue L, Pei X X, Kong F J, Zhao L, Lin X Y. 2023. Divergence of functions and expression patterns of soybean bZIP transcription factors. Front Plant Sci, 14: 1150363. |
| [36] | Zhang C Y, Fang H, Shi X T, He F, Wang R Y, Fan J B, Bai P F, Wang J Y, Park C H, Bellizzi M, Zhou X P, Wang G L, Ning Y S. 2020. A fungal effector and a rice NLR protein have antagonistic effects on a Bowman-Birk trypsin inhibitor. Plant Biotechnol J, 18(11): 2354-2363. |
| [37] | Zhang F, Fang H, Wang M, He F, Tao H, Wang R Y, Long J W, Wang J Y, Wang G L, Ning Y S. 2022. APIP5 functions as a transcription factor and an RNA-binding protein to modulate cell death and immunity in rice. Nucleic Acids Res, 50(9): 5064-5079. |
| [38] | Zhao K, Chen S, Yao W J, Cheng Z H, Zhou B R, Jiang T B. 2021. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol, 21(1): 122. |
| [39] | Zong W, Yang J, Fu J, Xiong L Z. 2020. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. J Integr Plant Biol, 62(6): 723-729. |
| [1] | M. Iqbal R. Khan, Sarika Kumari, Faroza Nazir, Risheek Rahul Khanna, Ravi Gupta, Himanshu Chhillar. Defensive Role of Plant Hormones in Advancing Abiotic Stress-Resistant Rice Plants [J]. Rice Science, 2023, 30(1): 15-35. |
| [2] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [3] | Chuxin Wang, Chengchao Zhu, Yu Zhou, Min Xiong, Jindong Wang, Huang Bai, Chenya Lu, Changquan Zhang, Qiaoquan Liu, Qianfeng Li. OsbZIP09, a Unique OsbZIP Transcription Factor of Rice, Promotes Rather Than Suppresses Seed Germination by Attenuating Abscisic Acid Pathway [J]. Rice Science, 2021, 28(4): 358-367. |
| [4] | Yongqi He, Jia Zhao, Defeng Feng, Zhibo Huang, Jiaming Liang, Yufei Zheng, Jinping Cheng, Jifeng Ying, Zhoufei Wang. RNA-Seq Study Reveals AP2-Domain-Containing Signalling Regulators Involved in Initial Imbibition of Seed Germination in Rice [J]. Rice Science, 2020, 27(4): 302-314. |
| [5] | Xixu Peng, Ting Xiao, Jiao Meng, Zong Tao, Dinggang Zhou, Xinke Tang, Haihua Wang. Differential Expression of Rice Valine-Qlutamine Gene Family in Response to Nitric Oxide and Regulatory Circuit of OsVQ7 and OsWRKY24 [J]. Rice Science, 2020, 27(1): 10-20. |
| [6] | Bo Wang, Zhaohui Zhong, Huanhuan Zhang, Xia Wang, Binglin Liu, Lijia Yang, Xiangyan Han, Deshui Yu, Xuelian Zheng, Chunguo Wang, Wenqin Song, Chengbin Chen, Yong Zhang. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice [J]. Rice Science, 2019, 26(2): 98-108. |
| [7] | Jini D., Joseph B.. Physiological Mechanism of Salicylic Acid for Alleviation of Salt Stress in Rice [J]. Rice Science, 2017, 24(2): 97-108. |
| [8] | Ashwini Narasimha, Sivarajan Sajeevan Radha, Udayakumar Makarla, Nalkur Nataraja Karaba. Identification and Characterization of OsWRKY72 Variant in Indica Genotypes [J]. Rice Science, 2016, 23(6): 297-305. |
| [9] | AI Li-ping, SHEN Ao, GAO Zhi-chao, LI Zheng-long, SUN Qiong-lin, LI Ying-ying, LUAN Wei-jiang. Reverse Genetic Analysis of Transcription Factor OsHox9, a Member of Homeobox Family, in Rice [J]. RICE SCIENCE, 2014, 21(6): 312-317. |
| [10] | Neerja SOOD, B. S. SOHAL, J. S. LORE . Foliar Application of Benzothiadiazole and Salicylic Acid to Combat Sheath Blight Disease of Rice [J]. RICE SCIENCE, 2013, 20(5): 349-355. |
| [11] | ZHAO Ming-hui, ZHANG Wen-zhong, MA Dian-rong, XU Zheng-jin, WANG Jia-yu, ZHANG Li, CHEN Wen-fu. Altered Expression of Transcription Factor Genes in Rice Flag Leaf under Low Nitrogen Stress [J]. RICE SCIENCE, 2012, 19(2): 100-107. |
| [12] | XU Gao-feng, ZHANG Fu-dou, LI Tian-lin, WU Di, ZHANG Yu-hua. Induced Effects of Exogenous Phenolic Acids on Allelopathy of a Wild Rice Accession (Oryza longistaminata, S37) [J]. RICE SCIENCE, 2010, 17(2): 134-140 . |
| [13] | SONG Yu, AI Chong-rui, JING Shao-juan, YU Di-qiu. Research Progress on Functional Analysis of Rice WRKY Genes [J]. RICE SCIENCE, 2010, 17(1): 60-72 . |
| [14] | MA Hao-li, ZHOU Han-lin, ZHANG Huai-yu, ZHAO Jie. Cloning and Expression Analysis of an AP2/ERF Gene and Its Responses to Phytohormones and Abiotic Stresses in Rice [J]. RICE SCIENCE, 2010, 17(1): 1-9 . |
| [15] | XIANG Dian-jun, HU Xiang-yang, ZHANG Yu, YIN Kui-de. Over-Expression of ICE1 Gene in Transgenic Rice Improves Cold Tolerance [J]. RICE SCIENCE, 2008, 15(3): 173-178 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||