
Rice Science ›› 2024, Vol. 31 ›› Issue (2): 203-214.DOI: 10.1016/j.rsci.2023.11.008
• Research Papers • Previous Articles Next Articles
Xie Shuwei, Shi Huanbin, Wen Hui, Liu Zhiquan, Qiu Jiehua, Jiang Nan( ), Kou Yanjun(
), Kou Yanjun( )
)
Received:2023-08-29
Accepted:2023-11-17
Online:2024-03-28
Published:2024-04-11
Contact:
Kou Yanjun (Xie Shuwei, Shi Huanbin, Wen Hui, Liu Zhiquan, Qiu Jiehua, Jiang Nan, Kou Yanjun. Carbon Catabolite Repressor UvCreA is Required for Development and Pathogenicity in Ustilaginoidea virens[J]. Rice Science, 2024, 31(2): 203-214.
Add to citation manager EndNote|Ris|BibTeX
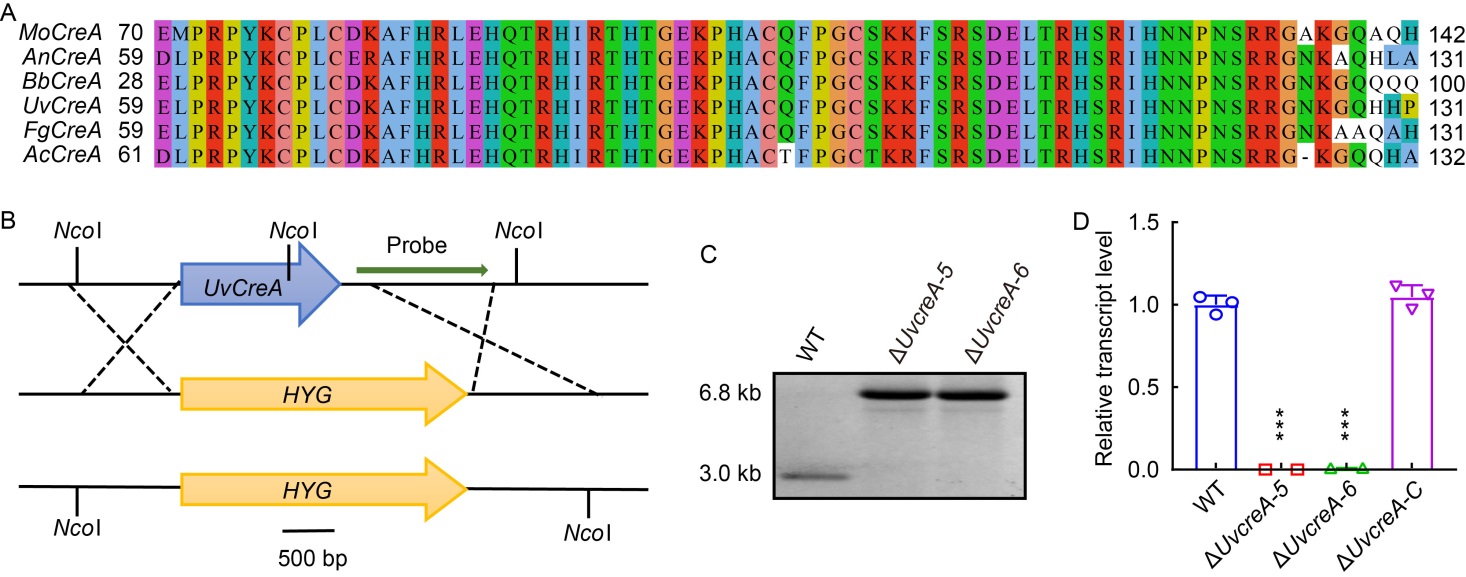
Fig. 1. Identification and targeted knockout of UvCreA in Ustilaginoidea virens. A, Functional domain analysis of CreA homologs from Magnaporthe oryzae (XP_003714427.1), Aspergillus nidulans (XP_663799.1), Beauveria bassiana (KAH8718674.1), U. virens (XP_042997845.1), Fusarium graminearum (XP_011327987.1), and Alternaria citri (BAF74640.1). B, Map of UvCreA knockout strategy and sites for restriction enzyme NcoI. HYG, Hygromycin resistance gene cassette.C, Southern blot analysis of UvCreA knockout mutants. The genomic DNA of the indicated strains was digested by NcoI and subjectedcom to Southern blotting with a probe located downstream of the UvCreA coding region. In the correct UvCreA knockout mutants, the 3.0 kb band in wild type (WT) was shifted to 6.8 kb. D, Expression level of UvCreA was determined by qRT-PCR assay. The UvCreA expression was detected in the WT and complementation (ΔUvcreA-C) strains but not in the ΔUvcreA-5 and ΔUvcreA-6 mutants. Data are Mean ± SD (n = 3). Asterisks (***) represent statistically significant differences at P ˂ 0.001.
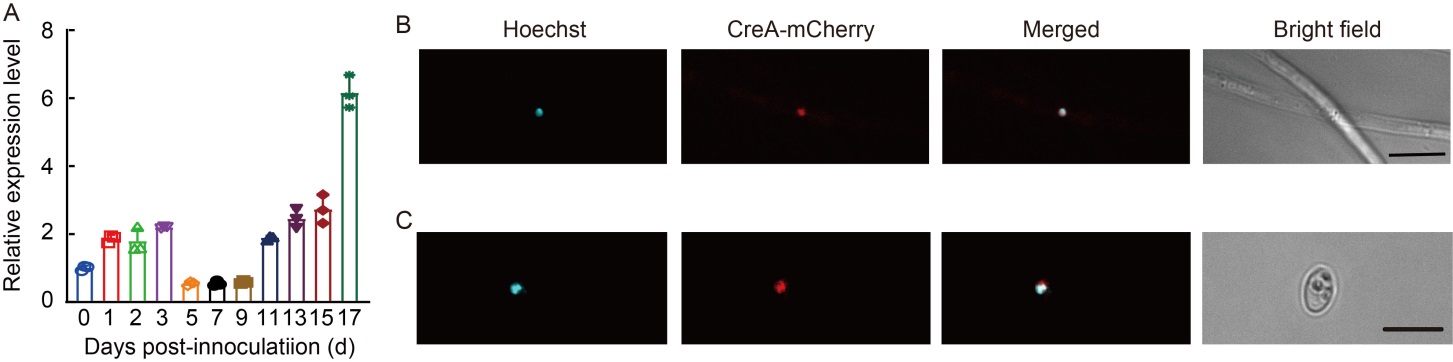
Fig. 2. Expression pattern and subcellular location of UvCreA. A, Relative expression levels of UvCreA gene during infection stages in Ustilaginoidea virens. With the β-tubulin gene as an internal control, the expression level of UvCreA at the vegetative growth stage was used as a criterion. Data are Mean ± SD (n = 3).B and C, UvCreA-mCherry co-localized with Hoechst-stained nuclei on hyphae (B) and conidia (C). Hoechst is used for staining nuclei in cells. Scale bars, 10 μm.
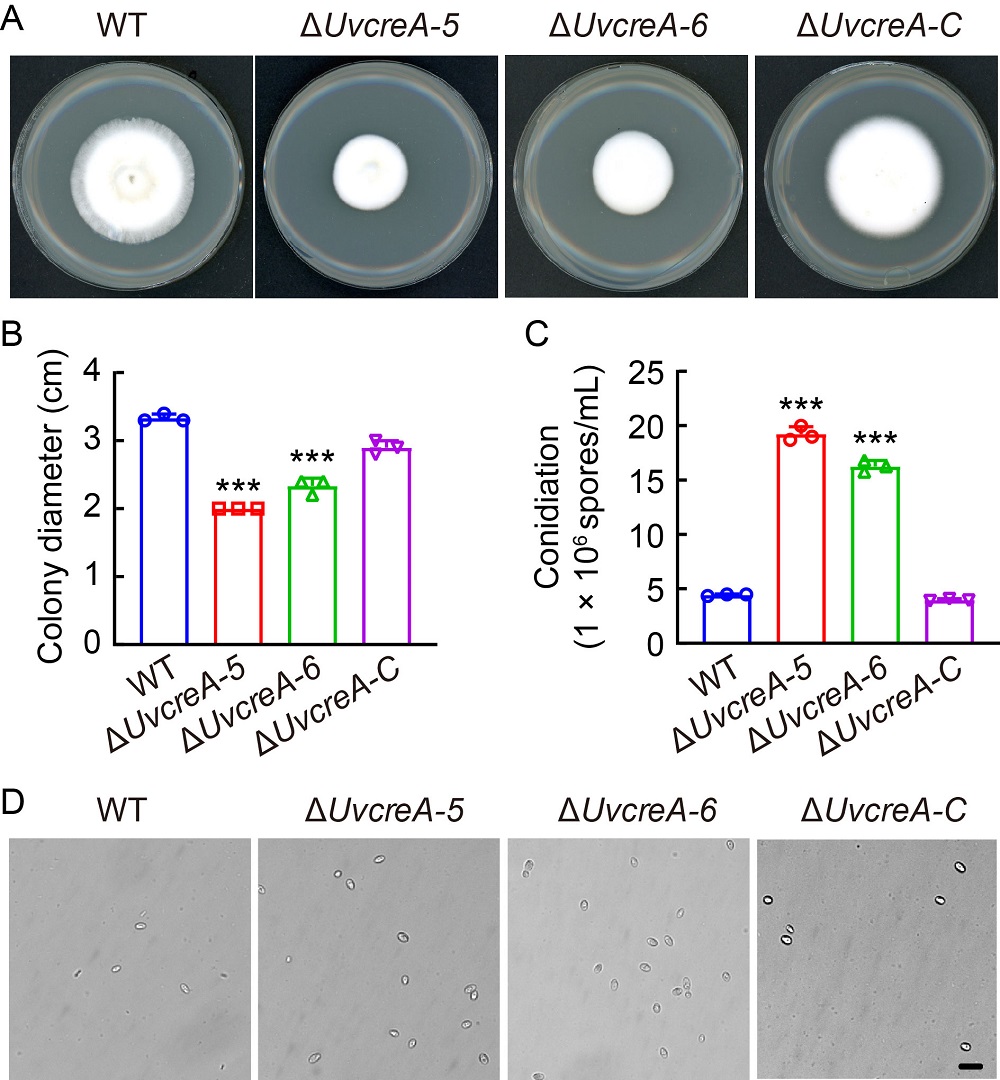
Fig. 3. Deletion of UvCreA resulted in decreased vegetative growth and increased spore production in Ustilaginoidea virens. A, Mycelium growths of wild type (WT), ΔUvcreA-5, ΔUvcreA-6, and complementation (ΔUvcreA-C) strains were measured after 14 d of cultivating on potato sucrose agar medium at 28 ºC. B, Colony diameters of WT, ΔUvcreA-5, ΔUvcreA-6, and ΔUvcreA-C strains. C and D, Deletion of UvCreA increased spore production in liquid potato sucrose medium for 7 d. Scale bar, 2.5 μm.Data are Mena ± SD (n = 3). Asterisks (***) represent statistically significant differences at P ˂ 0.001.
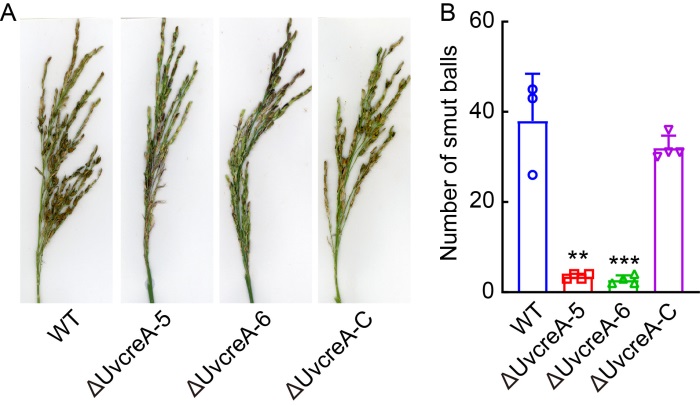
Fig. 4. Disruption of UvCreA gene resulted in reduced pathogenicity. A, Infection assays of wild type (WT), ΔUvcreA-5, ΔUvcreA-6, and complementation (ΔUvcreA-C) strains. At the booting stage, the panicles of Wanxian 98 were injected with spores and mycelia from the WT, ΔUvcreA-5, ΔUvcreA-6, and ΔUvcreA-C strains. After 3 weeks, the symptoms were determined.B, ΔUvcreA-5 and ΔUvcreA-6 strains exhibited a lower number of rice false smut balls compared with the WT and ΔUvcreA-C strains. Data are Mena ± SD (n = 4). Asterisks indicate statistically significant differences at P ˂ 0.01 (**) or P ˂ 0.001 (***).
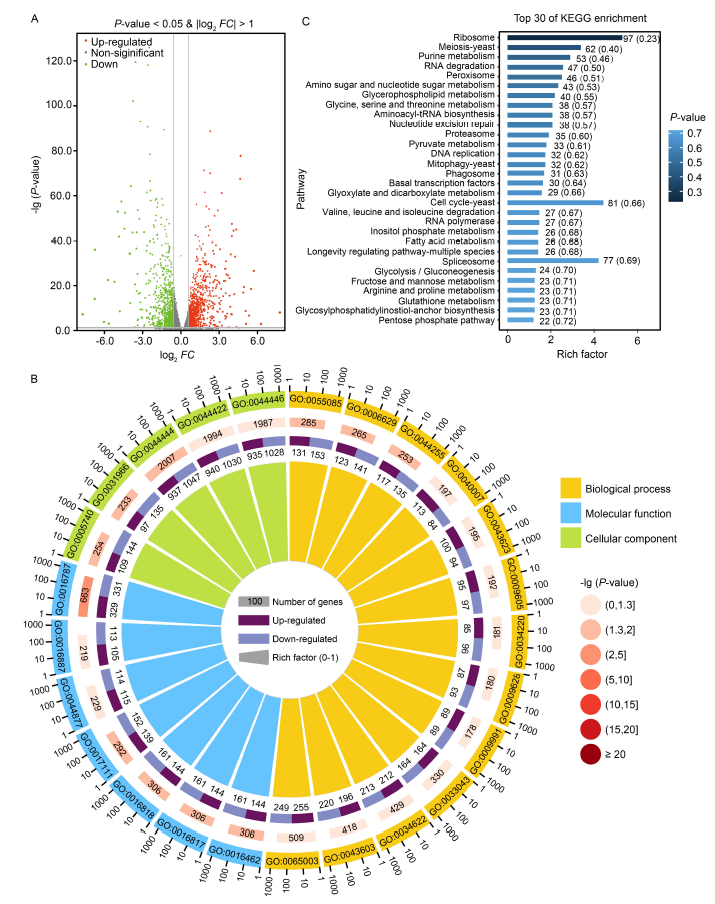
Fig. 5. RNA-seq analysis revealed effects of UvCreA deletion on transcription in Ustilaginoidea virens. A, Differentially expressed genes (DEGs) between ΔUvcreA and the wild type are depicted on a volcano plot. The x-axis represents the fold changes (FC) of up-regulated or down-regulated DEGs, and the y-axis indicates the levels of statistical significance. The P-value signifies the degree of change in gene expression. Blue, gray, and red dots correspond to down-regulated genes, genes with no significant change, and up-regulated genes, respectively.B, The enriched Gene Ontology (GO) classifications for DEGs were depicted in a doughnut chart. These classifications were categorized into three broader processes: biological process, molecular function, and cellular component.C, Encyclopedia of Genes and Genomes (KEGG) enrichment of the top 30 in the comparison of ΔUvcreA and wild type in U. virens.
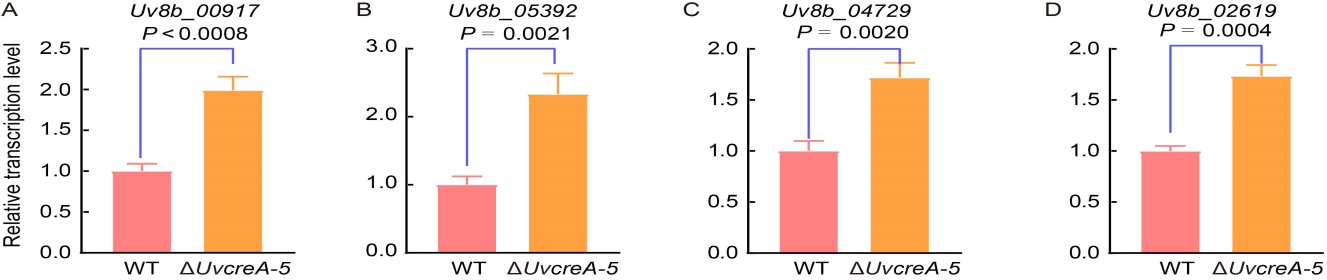
Fig. 6. UvCreA regulates transcription of glucose-repressed genes. A‒D, Relative transcription levels of xylanolytic gene (Uv8b_00917) (A), d-galactose utilization gene (Uv8b_05392) (B), maltose utilization gene (Uv8b_04729) (C), and galacturonic acid utilization gene (Uv8b_02619) (D) were up-regulated in the ΔUvcreA mutant compared with the wild type (WT) strain. Total mRNA was extracted from the mycelia of both the WT and ΔUvcreA mutant strains cultured in potato sucrose media after 7 d, with the β-tubulin gene serving as an internal control.
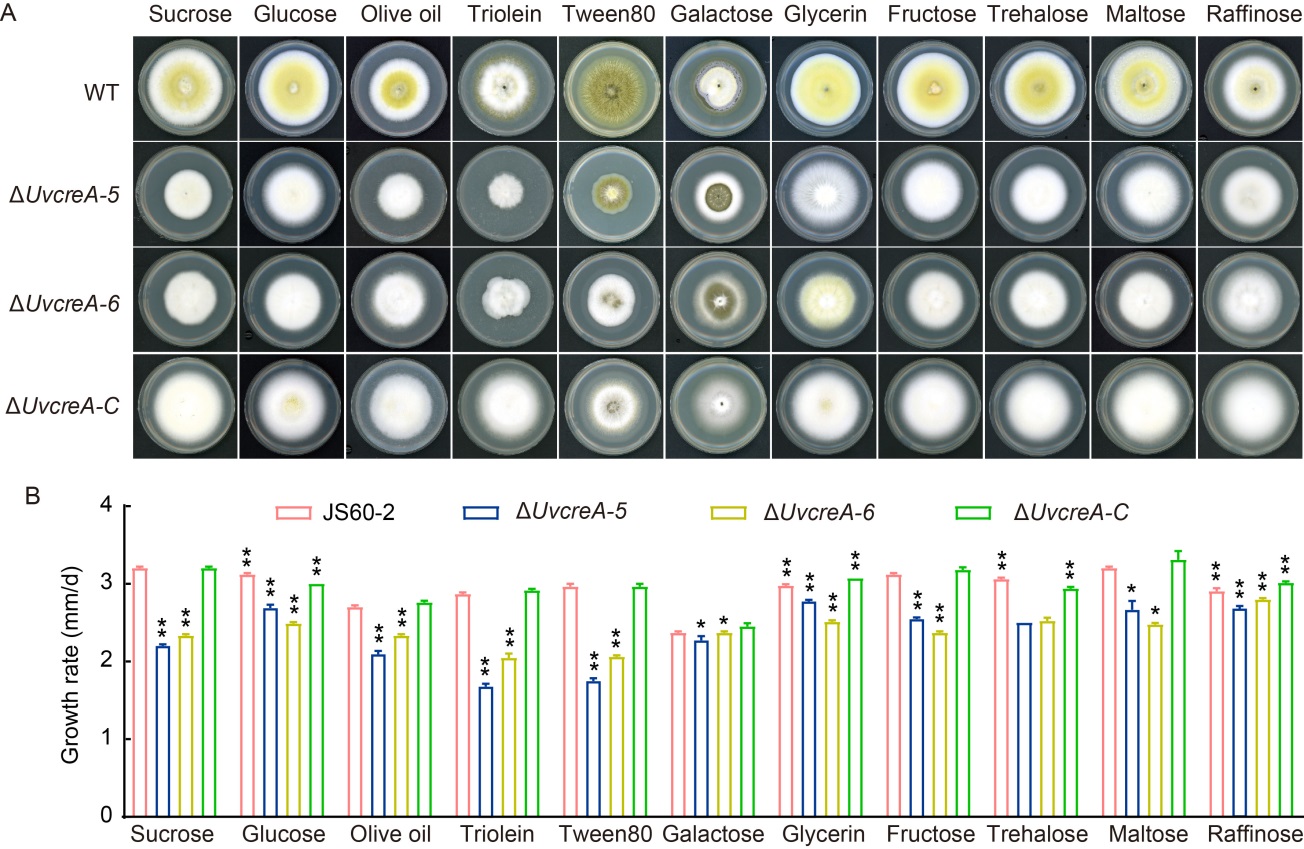
Fig. 7. UvCreA is required for utilization of various carbon sources. A, Strains grew on potato agar medium supplemented by various carbon sources at 28 ºC for 14 d.B, Growth rates of different strains were assessed at 14 d post-inoculation. Data are Mean ± SD (n = 3). Asterisks represent statistically significant differences at P ˂ 0.01 (**) and P ˂ 0.05 (*).
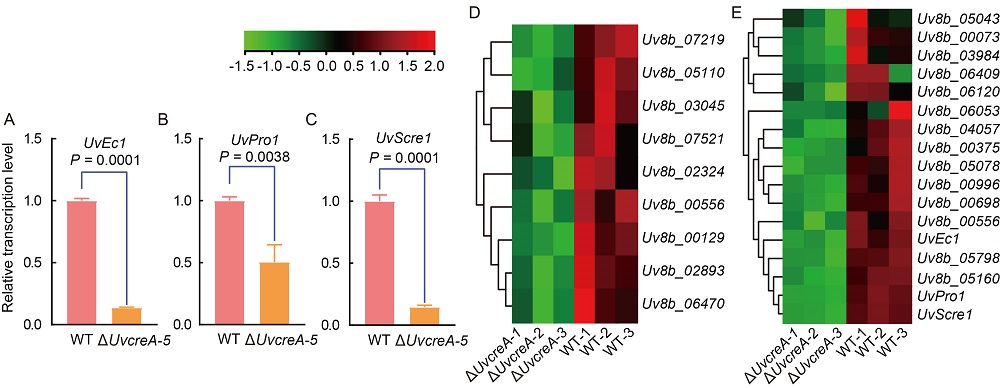
Fig. 8. UvCreA regulates transcription of genes involved in mitogen-activated protein kinase (MAPK) signaling pathway and pathogenesis. A‒C, Relative transcription levels of three identified pathogenic genes, including UvEc1 (A), UvPro1 (B), and UvScre1 (C), were down-regulated in the ΔUvcreA compared with the wild type (WT). Total mRNA was extracted from the mycelia of both the WT and ΔUvcreA mutant strains cultured in potato sucrose media after 7 d, with the β-tubulin gene serving as an internal control. Data are Mean ± SD (n = 3). D, The heatmap demonstrates the expression patterns of genes associated with the MAPK signaling pathway in the compared group, relative to the WT group.E, The heatmap illustrates the dynamic expression of pathogenic genes in the ΔUvcreA mutant in comparison with the WT strain. The colored cells in the images of D and E represent the detected expression levels of genes via RNA-seq.
| [1] | Adnan M, Zheng W H, Islam W, Arif M, Abubakar Y S, Wang Z H, Lu G D. 2017. Carbon catabolite repression in filamentous fungi. Int J Mol Sci, 19(1): 48. |
| [2] | Ahuatzi D, Riera A, Pelaez R, Herrero P, Moreno F. 2007. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem, 282(7): 4485-4493. |
| [3] | Alazi E, Knetsch T, Di Falco M, Reid I D, Arentshorst M, Visser J, Tsang A, Ram A F J. 2018. Inducer-independent production of pectinases in Aspergillus niger by overexpression of the D- galacturonic acid-responsive transcription factor gaaR. Appl Microbiol Biotechnol, 102(6): 2723-2736. |
| [4] | Beattie S R, Mark K M K, Thammahong A, Ries L N A, Dhingra S, Caffrey-Carr A K, Cheng C, Black C C, Bowyer P, Bromley M J, Obar J J, Goldman G H, Cramer R A. 2017. Filamentous fungal carbon catabolite repression supports metabolic plasticity and stress responses essential for disease progression. PLoS Pathog, 13(4): e1006340. |
| [5] | Cai Y Y, Wang J Y, Wu X Y, Liang S, Zhu X M, Li L, Lu J P, Liu X H, Lin F C. 2022. MoOpy2 is essential for fungal development, pathogenicity, and autophagy in Magnaporthe oryzae. Environ Microbiol, 24(3): 1653-1671. |
| [6] | Chen X Y, Pei Z X, Li P P, Li X B, Duan Y H, Liu H, Chen X L, Zheng L, Luo C X, Huang J B. 2021. Quantitative proteomics analysis reveals the function of the putative ester cyclase UvEC1 in the pathogenicity of the rice false smut fungus Ustilaginoidea virens. Int J Mol Sci, 22(8): 4069. |
| [7] | Fan J, Guo X Y, Li L, Huang F, Sun W X, Li Y, Huang Y Y, Xu Y J, Shi J, Lei Y, Zheng A P, Wang W M. 2015. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes. J Integr Plant Biol, 57(6): 577-590. |
| [8] | Fasoyin O E, Wang B, Qiu M G, Han X Y, Chung K R, Wang S H. 2018. Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in Aspergillus flavus. Fungal Genet Biol, 115: 41-51. |
| [9] | Franzino T, Boubakri H, Cernava T, Abrouk D, Achouak W, Reverchon S, Nasser W, Haichar F E. 2022. Implications of carbon catabolite repression for plant-microbe interactions. Plant Commun, 3(2): 100272. |
| [10] | Gancedo J M. 1998. Yeast carbon catabolite repression. Microbiol Mol Biol Rev, 62(2): 334-361. |
| [11] | Gomi K. 2019. Regulatory mechanisms for amylolytic gene expression in the koji mold Aspergillus oryzae. Biosci Biotechnol Biochem, 83(8): 1385-1401. |
| [12] | Hamel L P, Nicole M C, Duplessis S, Ellis B E. 2012. Mitogen- activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. Plant Cell, 24(4): 1327-1351. |
| [13] | Hong Y H, Cai R L, Guo J Y, Zhong Z H, Bao J D, Wang Z H, Chen X F, Zhou J, Lu G D. 2021. Carbon catabolite repressor MoCreA is required for the asexual development and pathogenicity of the rice blast fungus. Fungal Genet Biol, 146: 103496. |
| [14] | Jin Q C, Li C Y, Li Y Z, Shang J J, Li D B, Chen B S, Dong H T. 2013. Complexity of roles and regulation of the PMK1-MAPK pathway in mycelium development, conidiation and appressorium formation in Magnaporthe oryzae. Gene Expr Patterns, 13(5/6): 133-141. |
| [15] | Jonkers W, Rep M. 2009. Mutation of CRE1 in Fusarium oxysporum reverts the pathogenicity defects of the FRP1 deletion mutant. Mol Microbiol, 74(5): 1100-1113. |
| [16] | Katoh H, Ohtani K, Yamamoto H, Akimitsu K. 2007. Overexpression of a gene encoding a catabolite repression element in Alternaria citri causes severe symptoms of black rot in Citrus fruit. Phytopathology, 97(5): 557-563. |
| [17] | Kayikci Ö, Nielsen J. 2015. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res, 15(6): fov068. |
| [18] | Klaubauf S, Narang H M, Post H, Zhou M M, Brunner K, Mach- Aigner A R, Mach R L, Heck A J R, Maarten Altelaar A F, de Vries R P. 2014. Similar is not the same: Differences in the function of the (hemi-) cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet Biol, 72: 73-81. |
| [19] | Kowalczyk J E, Gruben B S, Battaglia E, Wiebenga A, Majoor E, de Vries R P. 2015. Genetic interaction of Aspergillus nidulans galR, xlnR and araR in regulating D-galactose and L-arabinose release and catabolism gene expression. PLoS One, 10(11): e0143200. |
| [20] | Kubicek C P, Mikus M, Schuster A, Schmoll M, Seiboth B. 2009. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels, 2: 19. |
| [21] | Kumar S, Kumar J. 2015. Evaluation of yield losses and management practices of false smut in rice (Oryza sativa). Indian Phytopathol, 68(1): 45-49. |
| [22] | Ladhalakshmi D, Laha G S, Singh R, Karthikeyan A, Mangrauthia S K, Sundaram R M, Thukkaiyannan P, Viraktamath B C. 2012. Isolation and characterization of Ustilaginoidea virens and survey of false smut disease of rice in India. Phytoparasitica, 40(2): 171-176. |
| [23] | Li G T, Zhang X, Tian H, Choi Y E, Tao W A, Xu J R. 2017. MST50 is involved in multiple MAP kinase signaling pathways in Magnaporthe oryzae. Environ Microbiol, 19(5): 1959-1974. |
| [24] | Lv B, Zheng L, Liu H, Tang J T, Hsiang T, Huang J B. 2016. Use of random T-DNA mutagenesis in identification of gene UvPRO1, a regulator of conidiation, stress response, and virulence in Ustilaginoidea virens. Front Microbiol, 7: 2086. |
| [25] | Mach-Aigner A R, Omony J, Jovanovic B, van Boxtel A J B, de Graaff L H. 2012. d-Xylose concentration-dependent hydrolase expression profiles and the function of CreA and XlnR in Aspergillus niger. Appl Environ Microbiol, 78(9): 3145-3155. |
| [26] | Matar K A O, Chen X F, Chen D J, Anjago W M, Norvienyeku J, Lin Y H, Chen M L, Wang Z H, Ebbole D J, Lu G D. 2017. WD40-repeat protein MoCreC is essential for carbon repression and is involved in conidiation, growth and pathogenicity of Magnaporthe oryzae. Curr Genet, 63(4): 685-696. |
| [27] | Meng S, Xiong M, Jagernath J S, Wang C C, Qiu J H, Shi H B, Kou Y J. 2020. UvAtg8-mediated autophagy regulates fungal growth, stress responses, conidiation, and pathogenesis in Ustilaginoidea virens. Rice, 13(1): 56. |
| [28] | Meng S, Liu Z Q, Shi H B, Wu Z L, Qiu J H, Wen H, Lin F C, Tao Z, Luo C X, Kou Y J. 2021. UvKmt6-mediated H3K27 trimethylation is required for development, pathogenicity, and stress response in Ustilaginoidea virens. Virulence, 12(1): 2972-2988. |
| [29] | Meng S, Qiu J H, Xiong M, Liu Z Q, Jagernath J S, Lin F C, Shi H B, Kou Y J. 2022a. UvWhi2 is required for stress response and pathogenicity in Ustilaginoidea virens. Rice Sci, 29(1): 47-54. |
| [30] | Meng S, Shi H B, Lin C Y, Wu Z L, Lin F C, Tao Z, Kou Y J. 2022b. UvKmt2-mediated H3K4 trimethylation is required for pathogenicity and stress response in Ustilaginoidea virens. J. Fungi , 8( 6): 553. |
| [31] | Nair A, Sarma S J. 2021. The impact of carbon and nitrogen catabolite repression in microorganisms. Microbiol Res, 251: 126831. |
| [32] | Park G, Xue C Y, Zhao X H, Kim Y, Orbach M, Xu J R. 2006. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell, 18(10): 2822-2835. |
| [33] | Qiu J H, Meng S, Deng Y Z, Huang S W, Kou Y J. 2019. Ustilaginoidea virens: A fungus infects rice flower and threats world rice production. Rice Sci, 26(4): 199-206. |
| [34] | Ries L N A, Beattie S R, Espeso E A, Cramer R A, Goldman G H. 2016. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics, 203(1): 335-352. |
| [35] | Soontorngun N. 2017. Reprogramming of nonfermentative metabolism by stress-responsive transcription factors in the yeast Saccharomyces cerevisiae. Curr Genet, 63(1): 1-7. |
| [36] | Sun W B, Wang A L, Xu D, Wang W X, Meng J J, Dai J G, Liu Y, Lai D W, Zhou L G. 2017. New ustilaginoidins from rice false smut balls caused by Villosiclava virens and their phytotoxic and cytotoxic activities. J Agric Food Chem, 65(25): 5151-5160. |
| [37] | Sun W X, Fan J, Fang A F, Li Y J, Tariqjaveed M, Li D Y, Hu D W, Wang W M. 2020. Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu Rev Phytopathol, 58: 363-385. |
| [38] | Suzuki K, Tanaka M, Konno Y, Ichikawa T, Ichinose S, Hasegawa- Shiro S, Shintani T, Gomi K. 2015. Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae. Appl Microbiol Biotechnol, 99(4): 1805-1815. |
| [39] | Tamayo E N, Villanueva A, Hasper A A, de Graaff L H, Ramón D, Orejas M. 2008. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol, 45(6): 984-993. |
| [40] | Tang J T, Bai J, Chen X Y, Zheng L, Liu H, Huang J B. 2020. Two protein kinases UvPmk1 and UvCDC2 with significant functions in conidiation, stress response and pathogenicity of rice false smut fungus Ustilaginoidea virens. Curr Genet, 66(2): 409-420. |
| [41] | Tannous J, Kumar D, Sela N, Sionov E, Prusky D, Keller N P. 2018. Fungal attack and host defence pathways unveiled in near- avirulent interactions of Penicillium expansum creA mutants on apples. Mol Plant Pathol, 19(12): 2635-2650. |
| [42] | Vega M, Riera A, Fernández-Cid A, Herrero P, Moreno F. 2016. Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of Mig1 protein repressor complex. J Biol Chem, 291(14): 7267-7285. |
| [43] | Wang R J, Peng J B, Li Q X, Peng Y L. 2017. Phosphorylation- mediated regulatory networks in mycelia of Pyricularia oryzae revealed by phosphoproteomic analyses. Mol Cell Proteom, 16(9): 1669-1682. |
| [44] | Wei Q H, Cui D Z, Zheng B J, Zhao M. 2023. In vitro antifungal activity of dihydrochelerythrine and proteomic analysis in Ustilaginoidea virens. Rice Sci, 30(3): 257-266. |
| [45] | Weinhandl K, Winkler M, Glieder A, Camattari A. 2014. Carbon source dependent promoters in yeasts. Microb Cell Fact, 13: 5. |
| [46] | Yoav S, Salame T M, Feldman D, Levinson D, Ioelovich M, Morag E, Yarden O, Bayer E A, Hadar Y. 2018. Effects of Cre1modification in the white-rot fungus Pleurotus ostreatus PC9: Altering substrate preference during biological pretreatment. Biotechnol Biofuels, 11: 212. |
| [47] | Zhang L S, Lubbers R J M, Simon A, Stassen J H M, Ribera P R V, Viaud M, van Kan J A L. 2016. A novel Zn2Cys6 transcription factor BcGaaR regulates D-galacturonic acid utilization in Botrytis cinerea. Mol Microbiol, 100(2): 247-262. |
| [48] | Zhang N, Yang J Y, Fang A F, Wang J Y, Li D Y, Li Y J, Wang S Z, Cui F H, Yu J J, Liu Y F, Peng Y L, Sun W X. 2020. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region. Mol Plant Pathol, 21(4): 445-459. |
| [49] | Zhang Y, Zhang K, Fang A F, Han Y Q, Yang J, Xue M F, Bao J D, Hu D W, Zhou B, Sun X Y, Li S J, Wen M, Yao N, Ma L J, Liu Y F, Zhang M, Huang F, Luo C X, Zhou L G, Li J Q, Chen Z Y, Miao J K, Wang S, Lai J S, Xu J R, Hsiang T, Peng Y L, Sun W X. 2014. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun, 5: 3849. |
| [50] | Zhao X H, Kim Y, Park G, Xu J R. 2005. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell, 17(4): 1317-1329. |
| [51] | Zheng X H, Fang A F, Qiu S S, Zhao G S, Wang J Y, Wang S Z, Wei J J, Gao H, Yang J Y, Mou B H, Cui F H, Zhang J, Liu J, Sun W X. 2022. Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity. Plant Cell, 34(8): 3088-3109. |
| [1] | Xia Xiaodong, Zhang Xiaobo, Wang Zhonghao, Cheng Benyi, Sun Huifeng, Xu Xia, Gong Junyi, Yang Shihua, Wu Jianli, Shi Yongfeng, Xu Rugen. Mapping and Functional Analysis of LE Gene in a Lethal Etiolated Rice Mutant at Seedling Stage [J]. Rice Science, 2023, 30(6): 567-576. |
| [2] | Md. Dhin Islam, Adam H. Price, Paul D. Hallett. Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth [J]. Rice Science, 2023, 30(5): 459-472. |
| [3] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| [4] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [5] | Wei Qinghui, Cui Daizong, Zheng Baojiang, Zhao Min. In Vitro Antifungal Activity of Dihydrochelerythrine and Proteomic Analysis in Ustilaginoidea virens [J]. Rice Science, 2023, 30(3): 257-266. |
| [6] | Liu Yueran, Qu Jinsong, Wang Yufu, Yin Weixiao, Luo Chaoxi. bZIP Transcription Factor UvATF21 Mediates Vegetative Growth, Conidiation, Stress Tolerance and Is Required for Full Virulence of Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2023, 30(1): 50-57. |
| [7] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [8] | Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice [J]. Rice Science, 2022, 29(3): 237-246. |
| [9] | Meng Shuai, Qiu Jiehua, Xiong Meng, Liu Zhiquan, Jane Sadhna Jagernath, Lin Fucheng, Shi Huanbin, Kou Yanjun. UvWhi2 Is Required for Stress Response and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2022, 29(1): 47-54. |
| [10] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [11] | Junjie Yu, Mina Yu, Tianqiao Song, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Xiaole Yin, Dong Liang, Yongfeng Liu. UvSMEK1, a Suppressor of MEK Null, Regulates Pathogenicity, Conidiation and Conidial Germination in Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2021, 28(5): 457-465. |
| [12] | Meng Xiong, Shuai Meng, Jiehua Qiu, Huanbin Shi, Xiangling Shen, Yanjun Kou. Putative Phosphatase UvPsr1 Is Required for Mycelial Growth, Conidiation, Stress Response and Pathogenicity in Ustilaginonidea virens [J]. Rice Science, 2020, 27(6): 529-536. |
| [13] | Shuting Yuan, Chunjue Xu, Wei Yan, Zhenyi Chang, Xingwang Deng, Zhufeng Chen, Jianxin Wu, Xiaoyan Tang. Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis [J]. Rice Science, 2020, 27(4): 289-301. |
| [14] | Vijayaraghavareddy Preethi, Xinyou Yin, C. Struik Paul, Makarla Udayakumar, Sreeman Sheshshayee. Responses of Lowland, Upland and Aerobic Rice Genotypes to Water Limitation During Different Phases [J]. Rice Science, 2020, 27(4): 345-354. |
| [15] | Hussain Kashif, Yingxing Zhang, Anley Workie, Riaz Aamir, Abbas Adil, Hasanuzzaman Rani Md., Hong Wang, Xihong Shen, Liyong Cao, Shihua Cheng. Association Mapping of Quantitative Trait Loci for Grain Size in Introgression Line Derived from Oryza rufipogon [J]. Rice Science, 2020, 27(3): 246-254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||