
Rice Science ›› 2020, Vol. 27 ›› Issue (3): 246-254.DOI: 10.1016/j.rsci.2020.04.007
• Research Paper • Previous Articles
Hussain Kashif1,2, Yingxing Zhang1, Anley Workie3, Riaz Aamir1, Abbas Adil1, Hasanuzzaman Rani Md.1,4, Hong Wang1, Xihong Shen1, Liyong Cao1( ), Shihua Cheng1(
), Shihua Cheng1( )
)
Received:2019-03-08
Accepted:2019-06-04
Online:2020-05-28
Published:2020-01-17
Hussain Kashif, Yingxing Zhang, Anley Workie, Riaz Aamir, Abbas Adil, Hasanuzzaman Rani Md., Hong Wang, Xihong Shen, Liyong Cao, Shihua Cheng. Association Mapping of Quantitative Trait Loci for Grain Size in Introgression Line Derived from Oryza rufipogon[J]. Rice Science, 2020, 27(3): 246-254.
Add to citation manager EndNote|Ris|BibTeX
| Chromosome | No. of markers | Size (Mb) | Spacing (kb) |
|---|---|---|---|
| 1 | 1007 | 43.2 | 15.5 |
| 2 | 757 | 35.8 | 15.8 |
| 3 | 742 | 36.4 | 17.4 |
| 4 | 876 | 35.4 | 18.1 |
| 5 | 625 | 29.7 | 17.1 |
| 6 | 811 | 31.2 | 16.3 |
| 7 | 632 | 29.6 | 18.7 |
| 8 | 458 | 28.3 | 15.3 |
| 9 | 397 | 22.9 | 16.8 |
| 10 | 211 | 23.2 | 15.3 |
| 11 | 301 | 29.0 | 15.5 |
| 12 | 256 | 27.5 | 17.7 |
Table 1 Distributions of markers on 12 chromosomes.
| Chromosome | No. of markers | Size (Mb) | Spacing (kb) |
|---|---|---|---|
| 1 | 1007 | 43.2 | 15.5 |
| 2 | 757 | 35.8 | 15.8 |
| 3 | 742 | 36.4 | 17.4 |
| 4 | 876 | 35.4 | 18.1 |
| 5 | 625 | 29.7 | 17.1 |
| 6 | 811 | 31.2 | 16.3 |
| 7 | 632 | 29.6 | 18.7 |
| 8 | 458 | 28.3 | 15.3 |
| 9 | 397 | 22.9 | 16.8 |
| 10 | 211 | 23.2 | 15.3 |
| 11 | 301 | 29.0 | 15.5 |
| 12 | 256 | 27.5 | 17.7 |
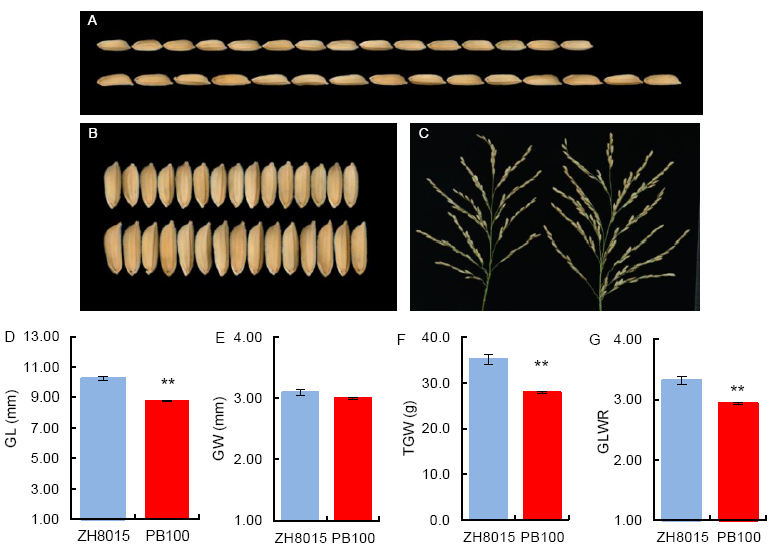
Fig. 2. Phenotypic comparisons of two parents for grain size related traits. A and D, Grain length (GL) of PB100 (Upper) and ZH8015 (Lower). B and E, Grain width (GW) of PB100 (Upper) and ZH8015 (Lower). C, Panicles of PB100 (Left) and ZH8015 (Right). F, 1000-grain weight (TGW). G, Grain length and width ratio (GLWR).Data are Mean ± SE (n = 3). The asterisks represent statistical significance between ZH8015 and PB100, determined by the Student’s t-test (**, P < 0.01).
| Trait | Environment | Mean ± SE | Range | H2B (%) | G × E |
|---|---|---|---|---|---|
| GL (mm) | Lingshui | 9.46 ± 0.04 | 8.60-10.40 | 56.3 | *** |
| Hangzhou | 9.61 ± 0.04 | 8.80-10.40 | |||
| GW (mm) | Lingshui | 3.42 ± 0.01 | 3.10-3.70 | 53.6 | *** |
| Hangzhou | 3.17 ± 0.02 | 2.90-3.80 | |||
| GLWR | Lingshui | 2.77 ± 0.01 | 2.50-3.30 | 49.2 | *** |
| Hangzhou | 3.05 ± 0.02 | 2.50-3.50 | |||
| TGW (g) | Lingshui | 36.40 ± 0.24 | 30.20-42.20 | 49.9 | *** |
| Hangzhou | 32.84 ± 0.22 | 26.90-40.30 |
Table 2 Phenotypic variations for grain traits across two environments.
| Trait | Environment | Mean ± SE | Range | H2B (%) | G × E |
|---|---|---|---|---|---|
| GL (mm) | Lingshui | 9.46 ± 0.04 | 8.60-10.40 | 56.3 | *** |
| Hangzhou | 9.61 ± 0.04 | 8.80-10.40 | |||
| GW (mm) | Lingshui | 3.42 ± 0.01 | 3.10-3.70 | 53.6 | *** |
| Hangzhou | 3.17 ± 0.02 | 2.90-3.80 | |||
| GLWR | Lingshui | 2.77 ± 0.01 | 2.50-3.30 | 49.2 | *** |
| Hangzhou | 3.05 ± 0.02 | 2.50-3.50 | |||
| TGW (g) | Lingshui | 36.40 ± 0.24 | 30.20-42.20 | 49.9 | *** |
| Hangzhou | 32.84 ± 0.22 | 26.90-40.30 |
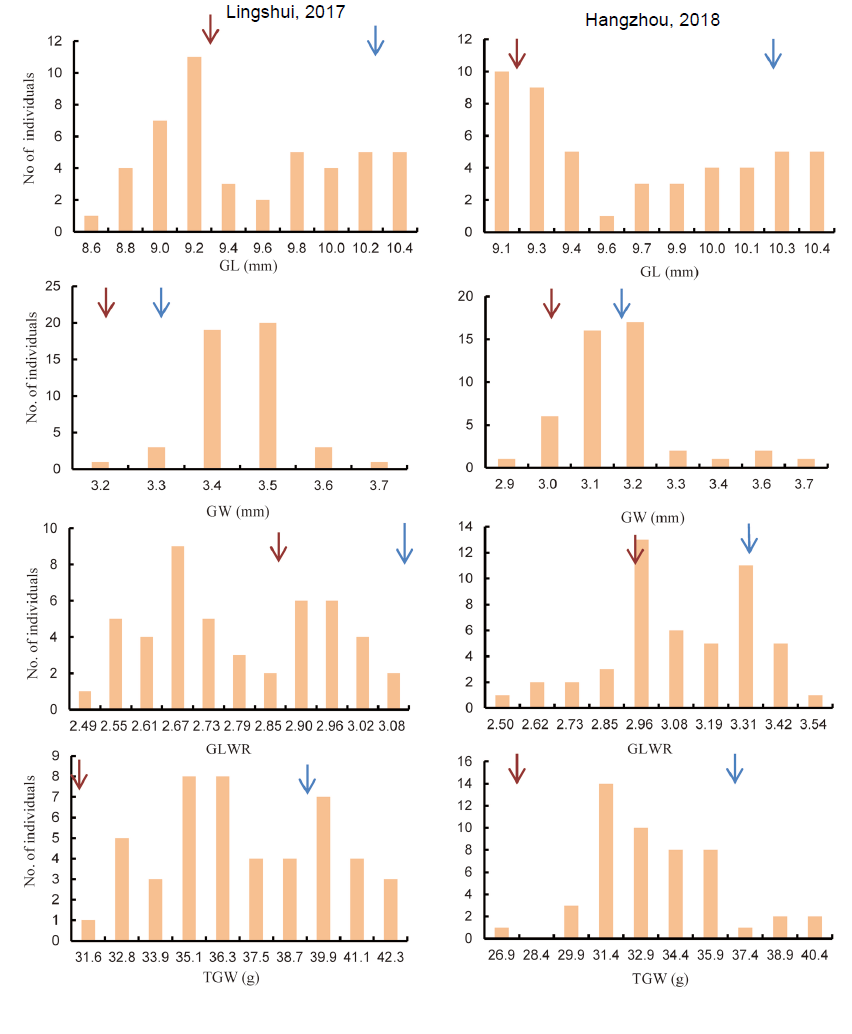
Supplemental Fig. 1. Frequency distribution and variation of grain shape related traits in introgression lines.GL, Grain length; GW, Grain width; GLWR, Grain length and grain width ratio; TGW, 1000-grain weight.Brown arrow indicate parent PB100 (small grains) and blue arrow indicate ZH8015 (Large grain).
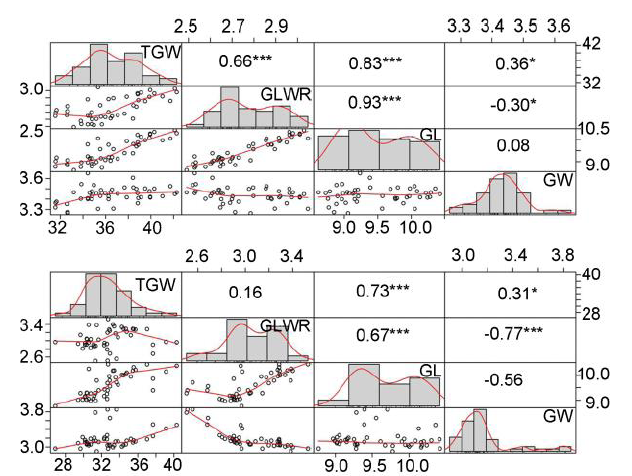
Fig. 3. Correlation analyses of four grain size traits.GL, Grain length; GW, Grain width; GLWR, Grain length-width ratio; TGW, 1000-grain weight. Top panel represent Lingshui-2017 and lower panel represent Hangzhou-2018. *, ** and *** represent the coefficient is significant at the 0.05, 0.01 and 0.001 levels, respectively.
| Trait | Position | Allele | MAF | P value | R2 (%) | Trait | Position | Allele | MAF | P value | R2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GL | 16 733 441 | A/G | 0.33 | 1.56E-15 | 45.74 | GLWR | 9 189 010 | T/C | 0.40 | 2.83E-11 | 44.24 |
| 16 734 121 | A/G | 0.33 | 3.29E-15 | 9 272 680 | A/C | 0.40 | 6.07E-11 | ||||
| 9 342 017 | A/T | 0.40 | 9.57E-15 | 25 042 825 | T/C | 0.40 | 6.07E-11 | ||||
| 9 228 066 | T/G | 0.39 | 1.52E-14 | 9 252 104 | A/C | 0.39 | 3.10E-10 | ||||
| 9 181 502 | T/C | 0.40 | 1.85E-14 | 25 043 314 | A/G | 0.27 | 8.60E-10 | ||||
| 9 529 975 | A/C | 0.39 | 2.85E-14 | 9 672 698 | A/G | 0.40 | 1.68E-09 | ||||
| 9 252 104 | A/C | 0.39 | 3.51E-13 | 9 270 291 | T/C | 0.39 | 6.81E-09 | ||||
| 9 254 860 | C/G | 0.40 | 1.04E-12 | 9 731 269 | A/T | 0.43 | 9.23E-09 | ||||
| 9 260 075 | T/G | 0.40 | 1.04E-12 | 9 776 599 | T/C | 0.34 | 6.67E-05 | ||||
| 9 189 010 | T/C | 0.40 | 1.06E-12 | TGW | 9 252 104 | A/C | 0.39 | 9.50E-10 | 63.00 | ||
| 9 372 896 | T/C | 0.40 | 4.11E-12 | 16 733 441 | A/G | 0.39 | 2.17E-09 | ||||
| 9 260 953 | T/C | 0.39 | 5.16E-12 | 16 734 121 | A/G | 0.09 | 2.39E-09 | ||||
| 25 043 314 | A/G | 0.27 | 1.57E-11 | 9 529 975 | A/C | 0.39 | 7.19E-09 | ||||
| 9 272 680 | A/G | 0.40 | 7.26E-11 | 9 342 017 | A/T | 0.40 | 3.40E-08 | ||||
| 25 042 825 | T/C | 0.40 | 9.73E-11 | 9 181 502 | T/C | 0.40 | 4.17E-08 | ||||
| 9 731 269 | A/T | 0.30 | 9.73E-11 | 9 228 066 | T/G | 0.39 | 4.18E-08 | ||||
| 9 270 029 | T/C | 0.43 | 1.15E-09 | 9 189 010 | T/C | 0.40 | 1.87E-07 | ||||
| 9 776 599 | T/C | 0.39 | 3.97E-09 | 9 254 860 | C/G | 0.40 | 3.54E-07 | ||||
| 9 342 017 | A/T | 0.30 | 1.71E-05 | 9 260 075 | T/G | 0.40 | 3.54E-07 | ||||
| GLWR | 16 733 441 | A/G | 0.09 | 9.73E-11 | 44.24 | 9 260 953 | T/C | 0.39 | 4.85E-07 | ||
| 16 734 121 | A/G | 0.39 | 2.44E-13 | 9 372 896 | T/C | 0.40 | 1.87E-06 | ||||
| 9 228 066 | T/G | 0.39 | 9.03E-13 | 25 043 314 | A/G | 0.27 | 3.68E-06 | ||||
| 9 181 502 | T/C | 0.40 | 9.30E-13 | 9 731 269 | A/T | 0.43 | 6.47E-06 | ||||
| 9 254 860 | C/G | 0.40 | 1.03E-12 | 9 672 698 | A/G | 0.40 | 1.02E-05 | ||||
| 9 260 075 | A/C | 0.40 | 1.03E-12 | 9 272 680 | A/G | 0.40 | 2.30E-05 | ||||
| 9 529 975 | A/C | 0.39 | 8.18E-12 | 25 042 825 | T/C | 0.40 | 2.30E-05 | ||||
| 9 372 896 | T/C | 0.40 | 8.72E-12 | 9 270 291 | T/C | 0.39 | 5.80E-05 | ||||
| 9 260 953 | T/C | 0.39 | 1.07E-11 |
Table 3 Fifty-six SNPs on rice chromosome 3 significantly associated with grain size in GWAS analyses.
| Trait | Position | Allele | MAF | P value | R2 (%) | Trait | Position | Allele | MAF | P value | R2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GL | 16 733 441 | A/G | 0.33 | 1.56E-15 | 45.74 | GLWR | 9 189 010 | T/C | 0.40 | 2.83E-11 | 44.24 |
| 16 734 121 | A/G | 0.33 | 3.29E-15 | 9 272 680 | A/C | 0.40 | 6.07E-11 | ||||
| 9 342 017 | A/T | 0.40 | 9.57E-15 | 25 042 825 | T/C | 0.40 | 6.07E-11 | ||||
| 9 228 066 | T/G | 0.39 | 1.52E-14 | 9 252 104 | A/C | 0.39 | 3.10E-10 | ||||
| 9 181 502 | T/C | 0.40 | 1.85E-14 | 25 043 314 | A/G | 0.27 | 8.60E-10 | ||||
| 9 529 975 | A/C | 0.39 | 2.85E-14 | 9 672 698 | A/G | 0.40 | 1.68E-09 | ||||
| 9 252 104 | A/C | 0.39 | 3.51E-13 | 9 270 291 | T/C | 0.39 | 6.81E-09 | ||||
| 9 254 860 | C/G | 0.40 | 1.04E-12 | 9 731 269 | A/T | 0.43 | 9.23E-09 | ||||
| 9 260 075 | T/G | 0.40 | 1.04E-12 | 9 776 599 | T/C | 0.34 | 6.67E-05 | ||||
| 9 189 010 | T/C | 0.40 | 1.06E-12 | TGW | 9 252 104 | A/C | 0.39 | 9.50E-10 | 63.00 | ||
| 9 372 896 | T/C | 0.40 | 4.11E-12 | 16 733 441 | A/G | 0.39 | 2.17E-09 | ||||
| 9 260 953 | T/C | 0.39 | 5.16E-12 | 16 734 121 | A/G | 0.09 | 2.39E-09 | ||||
| 25 043 314 | A/G | 0.27 | 1.57E-11 | 9 529 975 | A/C | 0.39 | 7.19E-09 | ||||
| 9 272 680 | A/G | 0.40 | 7.26E-11 | 9 342 017 | A/T | 0.40 | 3.40E-08 | ||||
| 25 042 825 | T/C | 0.40 | 9.73E-11 | 9 181 502 | T/C | 0.40 | 4.17E-08 | ||||
| 9 731 269 | A/T | 0.30 | 9.73E-11 | 9 228 066 | T/G | 0.39 | 4.18E-08 | ||||
| 9 270 029 | T/C | 0.43 | 1.15E-09 | 9 189 010 | T/C | 0.40 | 1.87E-07 | ||||
| 9 776 599 | T/C | 0.39 | 3.97E-09 | 9 254 860 | C/G | 0.40 | 3.54E-07 | ||||
| 9 342 017 | A/T | 0.30 | 1.71E-05 | 9 260 075 | T/G | 0.40 | 3.54E-07 | ||||
| GLWR | 16 733 441 | A/G | 0.09 | 9.73E-11 | 44.24 | 9 260 953 | T/C | 0.39 | 4.85E-07 | ||
| 16 734 121 | A/G | 0.39 | 2.44E-13 | 9 372 896 | T/C | 0.40 | 1.87E-06 | ||||
| 9 228 066 | T/G | 0.39 | 9.03E-13 | 25 043 314 | A/G | 0.27 | 3.68E-06 | ||||
| 9 181 502 | T/C | 0.40 | 9.30E-13 | 9 731 269 | A/T | 0.43 | 6.47E-06 | ||||
| 9 254 860 | C/G | 0.40 | 1.03E-12 | 9 672 698 | A/G | 0.40 | 1.02E-05 | ||||
| 9 260 075 | A/C | 0.40 | 1.03E-12 | 9 272 680 | A/G | 0.40 | 2.30E-05 | ||||
| 9 529 975 | A/C | 0.39 | 8.18E-12 | 25 042 825 | T/C | 0.40 | 2.30E-05 | ||||
| 9 372 896 | T/C | 0.40 | 8.72E-12 | 9 270 291 | T/C | 0.39 | 5.80E-05 | ||||
| 9 260 953 | T/C | 0.39 | 1.07E-11 |
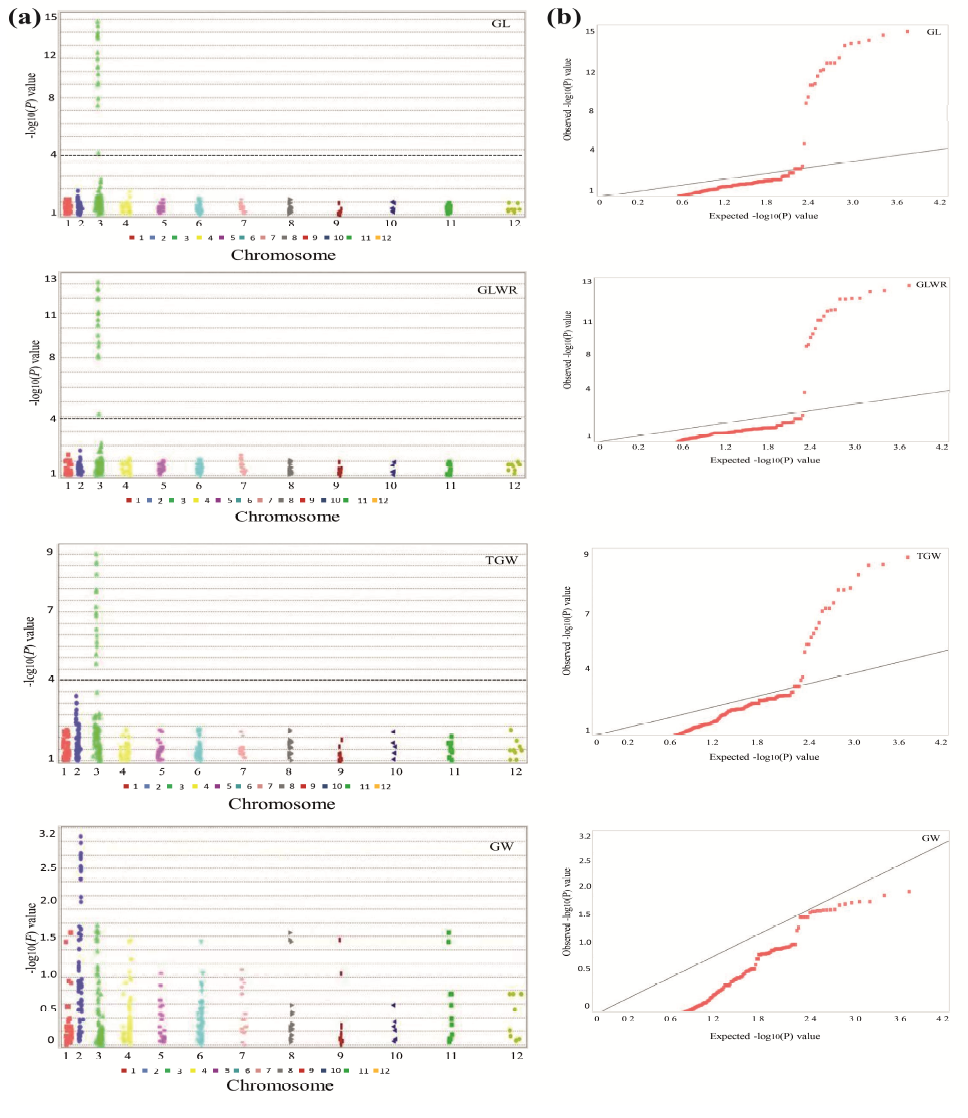
Supplemental Fig. 2. Manhattan plot (a) and Quantile-quantile plots (b) of four-grain shape related traits GL, Grain length; GW, Grain width; GLWR, Grain length-width ratio; TGW, Thousand grain weight.
| Candidate gene | SNP site | P value |
|---|---|---|
| Os03g0407400 | 16 733 441 | 1.56E-15 |
| Os03g0407400 | 16 734 121 | 3.29E-15 |
| Os03g0646900 | 25 042 825 | 9.73E-11 |
| Os03g0623700 | 25 043 314 | 1.71E-05 |
Table 4 Candidate genes/QTLs for grain length on chromosome 3.
| Candidate gene | SNP site | P value |
|---|---|---|
| Os03g0407400 | 16 733 441 | 1.56E-15 |
| Os03g0407400 | 16 734 121 | 3.29E-15 |
| Os03g0646900 | 25 042 825 | 9.73E-11 |
| Os03g0623700 | 25 043 314 | 1.71E-05 |
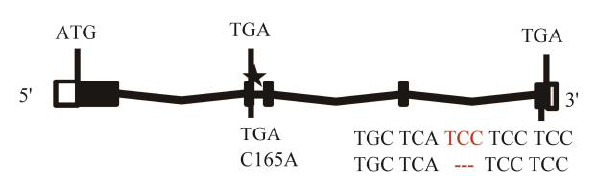
Fig. 5. Comparison and analyses of genomic DNA sequence of GS3.White boxes represent 5'- and 3'-UTR; black boxes indicate exons. Dark lines between exons represent introns. Translation start codon ATG and translation stop codon TGA indicated with the symbol of a star. C→A substitution shown in the second exon where C allele derived from small parent PB100 produce small grains and A allele derived from ZH8015 produce large grains, 3 bp TCC deletion found in all small grains indicated in exon 5.
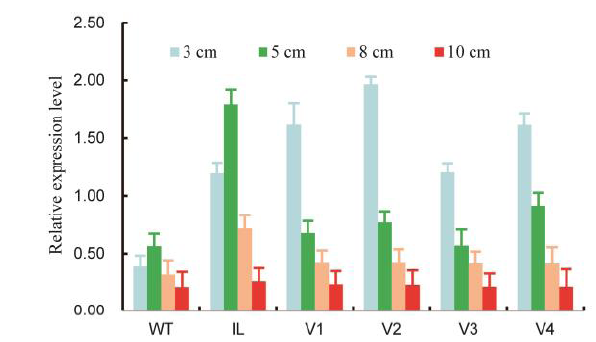
Fig. 6. Comparative expression pattern of GS3.WT, Wild type; V1, Changbai 25; V2, Zhejing 88; V3, Wuyunjing 27; V4, Zhejing 22. Comparative expression pattern of GS3 in introgression line (IL) and four CRISPR/Cas9-induced GS3 mutants using real-time RT-PCR analyses of young panicles at 3 cm, 5 cm, 8 cm and 10 cm in length.
| [1] | Agrama H A, Eizenga G C, Yan W G.2007. Association mapping of yield and its components in rice cultivars.Mol Breeding, 19(4): 341-356. |
| [2] | Bai X F, Luo L J, Yan W H, Kovi M R, Zhan W, Xing Y Z.2010. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locusqGL7. BMC Genet, 11(1): 16. |
| [3] | Barrantes W, Fernández-del-Carmen A, López-Casado G, González Sánchez M Á, Fernández-Muñoz R, Granell A, Monforte A J.2014. Highly efficient genomics-assisted development of a library of introgression lines ofSolanum pimpinellifolium. Mol Breeding, 34(4): 1817-1831. |
| [4] | Bradbury P J, Zhang Z W, Kroon D E, Casstevens T M, Ramdoss Y, Buckler E S.2007. TASSEL: Software for association mapping of complex traits in diverse samples.Bioinformatics, 23(19): 2633-2635. |
| [5] | Breseghello F, Sorrells M E.2006. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics, 172(2): 1165-1177. |
| [6] | Camus-Kulandaivelu L, Veyrieras J B, Madur D, Combes V, Fourmann M, Barraud S, Dubreuil P, Gouesnard B, Manicacci D, Charcosset A.2006. Maize adaptation to temperate climate: Relationship with population structure and polymorphism in theDwarf8 gene. Genetics, 172(4): 2449-2463. |
| [7] | Collard B C Y, Vera Cruz C M, McNally K L, Virk P S, Mackill D J. 2008. Rice molecular breeding laboratories in the genomics era: Current status and future considerations.Inter J Plant Genom, 2008: 524847. |
| [8] | Edzesi W M, Dang X J, Liang L J, Liu E B, Zaid I U, Hong D L.2016. Genetic diversity and elite allele mining for grain traits in rice (Oryza sativa L.) by association mapping. Front Plant Sci, 7: 787. |
| [9] | Ehrenreich I M, Hanzawa Y, Chou L, Roe J L, Kover P X, Purugganan M D.2009. Candidate gene association mapping ofArabidopsis flowering time. Genetics, 183(1): 325-335. |
| [10] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet, 112(6): 1164-1171. |
| [11] | Fan C C, Yu S B, Wang C R, Xing Y Z.2009. A causal C-A mutation in the second exon ofGS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet, 118(3): 465-472. |
| [12] | Feng Y, Lu Q, Zhai R R, Zhang M C, Xu Q, Yang Y L, Wang S, Yuan X P, Yu H Y, Wang Y P, Wei X H.2016. Genome wide association mapping for grain shape traits inindica rice. Planta, 244(4): 819-830. |
| [13] | Fu F H, Wang F, Huang W J, Peng H P, Wu Y Y, Huang D J.1994. Genetic analysis on grain characters in hybrid rice.Acta Agron Sin, 20(1): 39-45. (in Chinese with English abstract) |
| [14] | Gao Z Q, Zhan X D, Liang Y S, Cheng S H, Cao L Y.2011. Progress on genetics of rice grain shape trait and its related gene mapping and cloning.Hereditas, 33(4): 314-321. |
| [15] | Guo L B, Ma L L, Jiang H, Zeng D L, Hu J, Wu L W, Gao Z Y, Zhang G H, Qian Q.2009. Genetic analysis and fine mapping of two genes for grain shape and weight in rice.J Integr Plant Biol, 51(1): 45-51. |
| [16] | Hu J, Wang Y X, Fang Y X, Zeng L J, Xu J, Yu H P, Shi Z Y, Pan J J, Zhang D, Kang S J, Zhu L, Dong G J, Guo L B, Zeng D L, Zhang G H, Xie L H, Xiong G S, Li J Y, Qian Q.2015. A rare allele ofGS2 enhances grain size and grain yield in rice. Mol Plant, 8(10): 1455-1465. |
| [17] | Huang X H, Wei X H, Sang T, Zhao Q, Feng Q, Zhao Y, Li C Y, Zhu C R, Lu T T, Zhang Z W, Li M, Fan D L, Guo Y L, Wang A H, Wang L, Deng L W, Li W J, Lu Y Q, Weng Q J, Liu K Y, Huang T, Zhou T Y, Jing Y F, Li W, Lin Z, Buckler W S, Qian Q, Zhang Q F, Li J Y, Han B.2010. Genome-wide association studies of 14 agronomic traits in rice landraces.Nat Genet, 42(11): 961-967. |
| [18] | Huang X H, Han B.2014. Natural variations and genome-wide association studies in crop plants.Ann Rev Plant Biol, 65: 531-551. |
| [19] | Jia G Q, Huang X H, Zhi H, Zhao Y, Zhao Q, Li W J, Chai Y, Yang L F, Liu K Y, Lu H Y, Zhu C R, Lu Y Q, Zhou C C, Fan D L, Weng Q J, Guo Y L, Huang T, Zhang L, Lu T T, Feng Q, Hao H F, Liu H K, Lu P, Zhang N, Li Y H, Guo E H, Wang S J, Wang S Y, Liu J R, Zhang W F, Chen G Q, Zhang B J, Li W, Wang Y F, Li H Q, Zhao B H, Li J Y, Diao X M, Han B.2013. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet, 45(8): 957-961. |
| [20] | Jun T H, Van K, Kim M Y, Lee S H, Walker D R.2008. Association analysis using SSR markers to find QTL for seed protein content in soybean.Euphytica, 162(2): 179-191. |
| [21] | Kang H M, Zaitlen N A, Wade C M, Kirby A, Heckerman D, Daly M J, Eskin E.2008. Efficient control of population structure in model organism association mapping.Genetics, 178(3): 1709-1723. |
| [22] | Kato T, Segami S, Toriyama M, Kono I, Ando T, Yano M, Kitano H, Miura K, Iwasaki Y.2011. Detection of QTLs for grain length from large grain rice (Oryza sativa L.). Breeding Sci, 61(3): 269-274. |
| [23] | Khush G S.2005. What it will take to feed 5.0 billion rice consumers in 2030.Plant Mol Biol, 59(1): 1-6. |
| [24] | Li M M, Xu L, Ren J F, Cao G L, Yu L Q, He H H, Han L Z, Koh H J.2010. Identification of quantitative trait loci for grain traits injaponica rice. Agric Sci China, 9(7): 929-936. |
| [25] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H.2011. Natural variation inGS5 plays an important role in regulating grain size and yield in rice. Nat Genet, 43(12): 1266-1269. |
| [26] | Lipka A E, Tian F, Wang Q S, Peiffer J, Li M, Bradbury P J, Gore M A, Buckler E S, Zhang Z W.2012. GAPIT: Genome association and prediction integrated tool.Bioinformatics, 28(18): 2397-2399. |
| [27] | Liu D L, Kang M H, Wang F, Liu W G, Fu C Y, Li J H, Zhu M S, Zeng X Q, Liao Y L, Liu Z R.2015. Mapping of the genetic determinant for grain size in rice using a recombinant inbred line (RIL) population generated from two eliteindica parents. Euphytica, 206(1): 159-173. |
| [28] | Mao H L, Sun S Y, Yao J L, Wang C R, Yu S B, Xu C G, Li X H, Zhang Q F.2010. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice.Proc Natl Acad Sci USA, 107: 19579-19584. |
| [29] | Murray M G, Thompson W F.1980. Rapid isolation of high molecular weight plant DNA.Nucl Acids Res, 8(19): 4321-4325. |
| [30] | Olsen K M, Halldorsdottir S S, Stinchcombe J R, Weinig C, Schmitt J, Purugganan M D.2004. Linkage disequilibrium mapping ofArabidopsis CRY2 flowering time alleles. Genetics, 167(3): 1361-1369. |
| [31] | Palaisa K A, Morgante M, Williams M, Rafalski A.2003. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci.Plant Cell, 15(8): 1795-1806. |
| [32] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L W, Gao J P, Lin H X.2012. The novel quantitative trait locusGL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res, 22(12): 1666-1680. |
| [33] | Qiu X J, Pang Y L, Yuan Z H, Xing D Y, Xu J L, Dingkuhn M, Li Z K, Ye G Y.2015. Genome-wide association study of grain appearance and milling quality in a worldwide collection ofindica rice germplasm. PLoS One, 10(12): e0145577. |
| [34] | Rui C Q, Zhao A C.1983. Genetic analysis of weight and shape of F1’s grains by diallel crossing method in hsien rice.Sci Agric Sin, (5): 14-20. (in Chinese with English abstract) |
| [35] | Shi C H, Shen Z T.1994. Analysis of genetic effects of grain traits inindica rice. J Zhejiang Agric Univ, 20(4): 405-410. (in Chinese with English abstract) |
| [36] | Shi C H, Shen Z T.1996. Additive and dominance correlation analysis of grain shape and yield traits in indica rice.Acta Agron Sin, 22(1): 36-42. (in Chinese with English abstract) |
| [37] | Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M.2008. Deletion in a gene associated with grain size increased yields during rice domestication.Nat Genet, 40(8): 1023-1028. |
| [38] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shangguan Y Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B.2016. OsSPL13 controls grain size in cultivated rice.Nat Genet, 47(3): 447-456. |
| [39] | Singh R, Singh A K, Sharma T R, Singh A, Singh N K.2012. Fine mapping of grain length QTLs on chromosomes 1 and 7 in Basmati rice (Oryza sativa L.). J Plant Biochem Biotechnol, 21(2): 157-166. |
| [40] | Sun L J, Li X J, Fu Y C, Zhu Z F, Tan L B, Liu F X, Sun X Y, Sun X W, Sun C Q.2013. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol, 55(10): 938-949. |
| [41] | Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, Kanamori H, Padhukasahasram B, Bustamante C, Yoshimura A, Doi K, McCouch S. 2009. Evolutionary history of GS3, a gene conferring grain size in rice. Genetics, 182(4): 1323-1334. |
| [42] | Wan X Y, Weng J F, Zhai H Q, Wang J K, Lei C L, Liu X L, Guo T, Jiang L, Su N, Wan J M.2008. QTL analysis for rice grain width and fine mapping of an identified QTL allelegw-5 in a recombination hotspot region on chromosome 5. Genetics, 179(4): 2239-2252. |
| [43] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.2012. Control of grain size, shape and quality byOsSPL16 in rice. Nat Genet, 44(8): 950-954. |
| [44] | Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D.2015. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality.Nat Genet, 47(8): 949-954. |
| [45] | Wilson L M, Whitt S R, Ibáñez A M, Rocheford T R, Goodman M M, Buckler E S.2004. Dissection of maize kernel composition and starch production by candidate gene association.Plant Cell, 16(10): 2719-2733. |
| [46] | Yang T F, Zeng R Z, Zhu H T, Chen L, Zhang Z M, Ding X H, Li W T, Zhang G Q.2010. Effect of grain length geneGS3 in pyramiding breeding of rice. Mol Plant Breeding, 8(1): 59-66. |
| [47] | Yang W N, Guo Z L, Huang C L, Duan L F, Chen G X, Jiang N, Fang W, Feng H, Xie W B, Lian X M, Wang G W, Luo Q M, Zhang Q F, Liu Q, Xiong L Z.2014. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice.Nat Commun, 5: 5087. |
| [48] | Yano K, Yamamoto E, Aya K, Takeuchi H, Lo P C, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, Matsuoka M.2016. Genome- wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice.Nat Genet, 48(8): 927-934. |
| [49] | Yu S W, Yang C D, Fan Y Y, Zhuang J Y, Li X M.2008. Genetic dissection of a thousand-grain weight quantitative trait locus on rice chromosome 1.Chin Sci Bull, 53(15): 2326-2332. |
| [50] | Zhang W H, Sun P Y, He Q, Shu F, Wang J, Deng H F.2013. Fine mapping ofGS2, a dominant gene for big grain rice. Crop J, 1(2): 160-165. |
| [51] | Zhang X J, Wang J F, Huang J, Lan H X, Wang C L, Yin C F, Wu Y Y, Tang H J, Qian Q, Li J Y, Zhang H S.2012. Rare allele ofOsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA, 109: 21534-21539. |
| [52] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q.2018. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat Commun, 9(1): 1240. |
| [53] | Zhao K Y, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R.2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits inOryza sativa. Nat Commun, 2: 467. |
| [54] | Zhou Q Y, An H, Zhang Y, Shen F C.2000. Study on heredity of morphological characters of rice grain.J Southwest Agric Univ, 22(2): 102-104. (in Chinese with English abstract) |
| [55] | Zhu C S, Gore M, Buckler E S, Yu J M.2008. Status and prospects of association mapping in plants.Plant Genom, 1(1): 5-20. |
| [1] | Md. Dhin Islam, Adam H. Price, Paul D. Hallett. Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth [J]. Rice Science, 2023, 30(5): 459-472. |
| [2] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [3] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [4] | Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice [J]. Rice Science, 2022, 29(3): 237-246. |
| [5] | Yan Wang, Xiaoqin Zeng, Lu Lu, Qinglan Cheng, Fayu Yang, Mingjiang Huang, Mao Xiong, Yunfeng Li. MULTI-FLORET SPIKELET 4 (MFS4) Regulates Spikelet Development and Grain Size in Rice [J]. Rice Science, 2021, 28(4): 344-357. |
| [6] | Yuyu Chen, Aike Zhu, Pao Xue, Xiaoxia Wen, Yongrun Cao, Beifang Wang, Yue Zhang, Liaqat Shah, Shihua Cheng, Liyong Cao, Yingxin Zhang. Effects of GS3 and GL3.1 for Grain Size Editing by CRISPR/Cas9 in Rice [J]. Rice Science, 2020, 27(5): 405-413. |
| [7] | Vijayaraghavareddy Preethi, Xinyou Yin, C. Struik Paul, Makarla Udayakumar, Sreeman Sheshshayee. Responses of Lowland, Upland and Aerobic Rice Genotypes to Water Limitation During Different Phases [J]. Rice Science, 2020, 27(4): 345-354. |
| [8] | Shuting Yuan, Chunjue Xu, Wei Yan, Zhenyi Chang, Xingwang Deng, Zhufeng Chen, Jianxin Wu, Xiaoyan Tang. Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis [J]. Rice Science, 2020, 27(4): 289-301. |
| [9] | Zhongkang Wang, Dongdong Zeng, Ran Qin, Jialin Liu, Chunhai Shi, Xiaoli Jin. A Novel and Pleiotropic Factor SLENDER GRAIN3 Is Involved in Regulating Grain Size in Rice [J]. Rice Science, 2018, 25(3): 132-141. |
| [10] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [11] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| [12] | Fernando Polesi Luís, Bruder Silveira Sarmento Silene, Guidolin Canniatti-Brazaca Solange. Starch Digestibility and Functional Properties of Rice Starch Subjected to Gamma Radiation [J]. Rice Science, 2018, 25(1): 42-51. |
| [13] | Singh Bhupinder, Raja Reddy Kambham, Diaz Redoña Edilberto, Walker Timothy. Screening of Rice Cultivars for Morpho-Physiological Responses to Early-Season Soil Moisture Stress [J]. Rice Science, 2017, 24(6): 322-335. |
| [14] | Rekha Talukdar Preeti, Rathi Sunayana, Pathak Khanin, Kumar Chetia Sanjay, Nath Sarma Ramendra. Population Structure and Marker-Trait Association in Indigenous Aromatic Rice [J]. Rice Science, 2017, 24(3): 145-154. |
| [15] | Jini D., Joseph B.. Physiological Mechanism of Salicylic Acid for Alleviation of Salt Stress in Rice [J]. Rice Science, 2017, 24(2): 97-108. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||