
Rice Science ›› 2025, Vol. 32 ›› Issue (5): 685-703.DOI: 10.1016/j.rsci.2025.05.004
• Research Papers • Previous Articles Next Articles
Mareyam Mukhtar1,2, Amresh Kumar1, Ashfak S. Mujawar1,2, Bhuvnesh Sareen1, Suhas G. Karkute1,3, Rohini Sreevathsa1, Amitha Mithra Sevanthi1, Amolkumar U. Solanke1( )
)
Received:2025-02-12
Accepted:2025-05-22
Online:2025-09-28
Published:2025-10-11
Contact:
Amolkumar U. Solanke (Mareyam Mukhtar, Amresh Kumar, Ashfak S. Mujawar, Bhuvnesh Sareen, Suhas G. Karkute, Rohini Sreevathsa, Amitha Mithra Sevanthi, Amolkumar U. Solanke. Genome-Wide Identification of Dopamine β-Monooxygenase N-Terminal Gene Family in Rice and Its Role in Response to Blast Disease and Abiotic Stress[J]. Rice Science, 2025, 32(5): 685-703.
Add to citation manager EndNote|Ris|BibTeX

Fig. 1. A comprehensive schematic illustrating genomic organization and structural features of OsDOMON genes. A, Schematic representation of the chromosomal distribution of rice DOMON genes. The physical positions (cM) of OsDOMONs were mapped according to the rice genome. B, Gene structures illustrating the exon-intron organization of the DOMON genes in rice. CDS, Coding sequence. C, Conserved domain analysis of OsDOMONs was conducted using TBtools. This analysis revealed the conservation of the OsDOMON domain along with other conserved domains like the Cytochrome_b_N superfamily among the genes. Different domains are represented in different colors as indicated in the figure. D, Conserved motifs of OsDOMON peptides were investigated using MEME. Different conserved motifs are represented by different colored boxes, and the motif sequences are given at the bottom. The size of the boxes indicates the length of the conserved motifs.
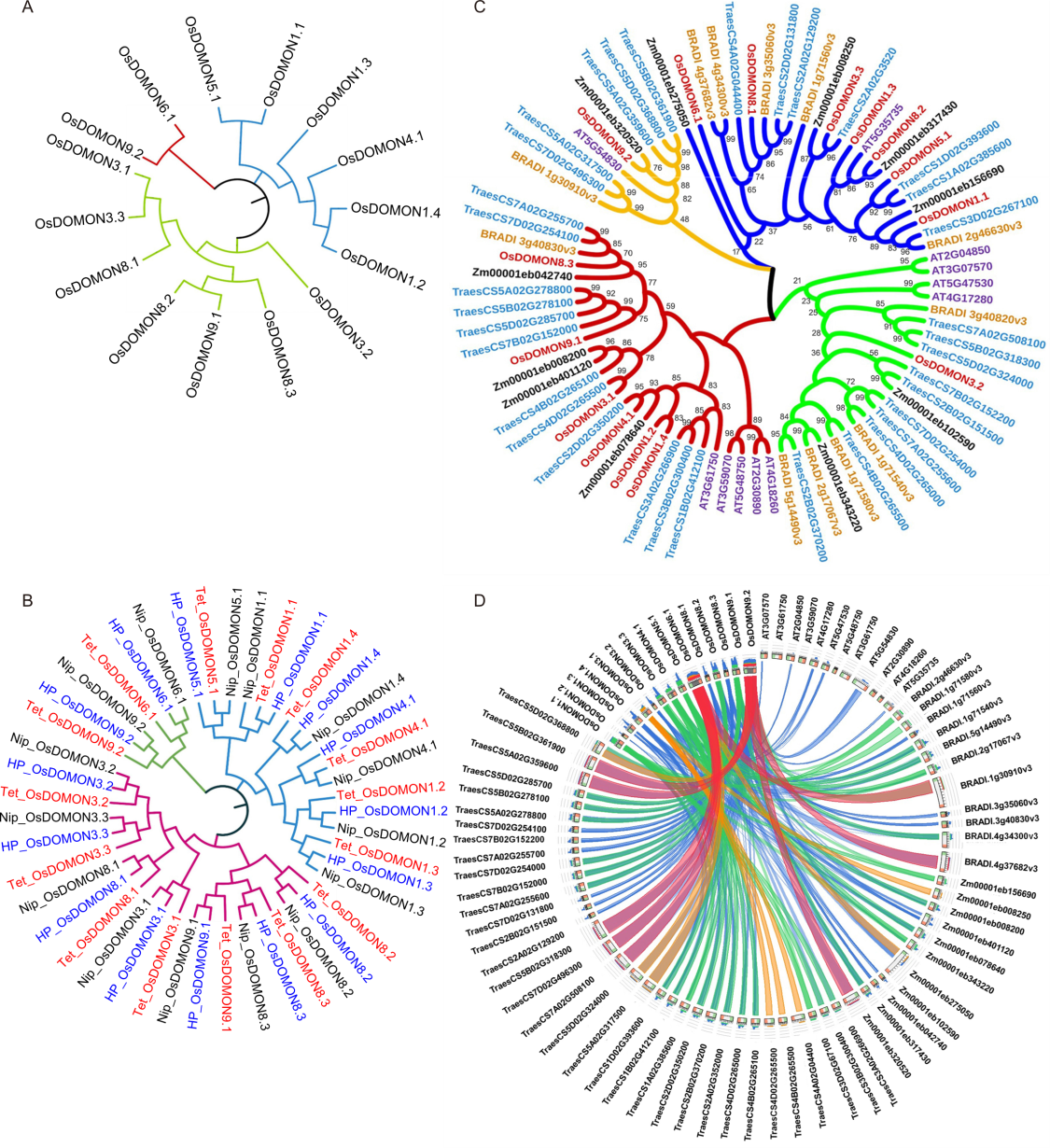
Fig. 2. Phylogenetic analysis and synteny analysis of DOMON proteins in rice and other plant species. A, Phylogenetic relationship among different OsDOMONs. B, Phylogenetic relationship between different OsDOMONs in rice from three different cultivars HP2216 (HP), Nipponbare (NIP), and Tetep (Tet). OsDOMONs from HP2216 are marked in blue, Nipponbare in black, and Tetep in red. The tree was constructed based on the full-length sequences of proteins. C, Phylogenetic relationships of DOMON proteins from five different plant species were generated by MEGAX with 1000 bootstrap replicates using the maximum likelihood method. The DOMON genes from O. sativa are marked in maroon, yellow for Brachypodium distachyon, blue for Triticum aestivum, purple for Arabidopsis thaliana and black for Zea mays. D, Visualization of evolutionary relationship of DOMONs from four different plant species B. distachyon, T. aestivum, A. thaliana, and Z. mays using Circos plot.
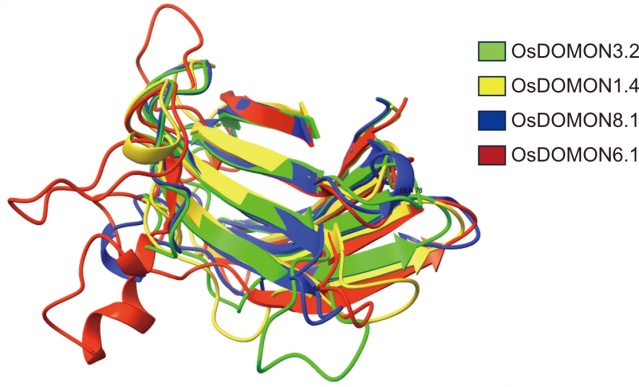
Fig. 3. Superimposed AlphaFold-predicted 3D structures of representative OsDOMON proteins. Structural alignment of OsDOMON3.2, OsDOMON1.4, OsDOMON6.1, and OsDOMON8.1 illustrates the conserved β-sandwich fold characteristic of DOMON domains. Despite differences in the adjacent domain architecture, the core structural fold is preserved across all four models, indicating strong structural conservation within the family.
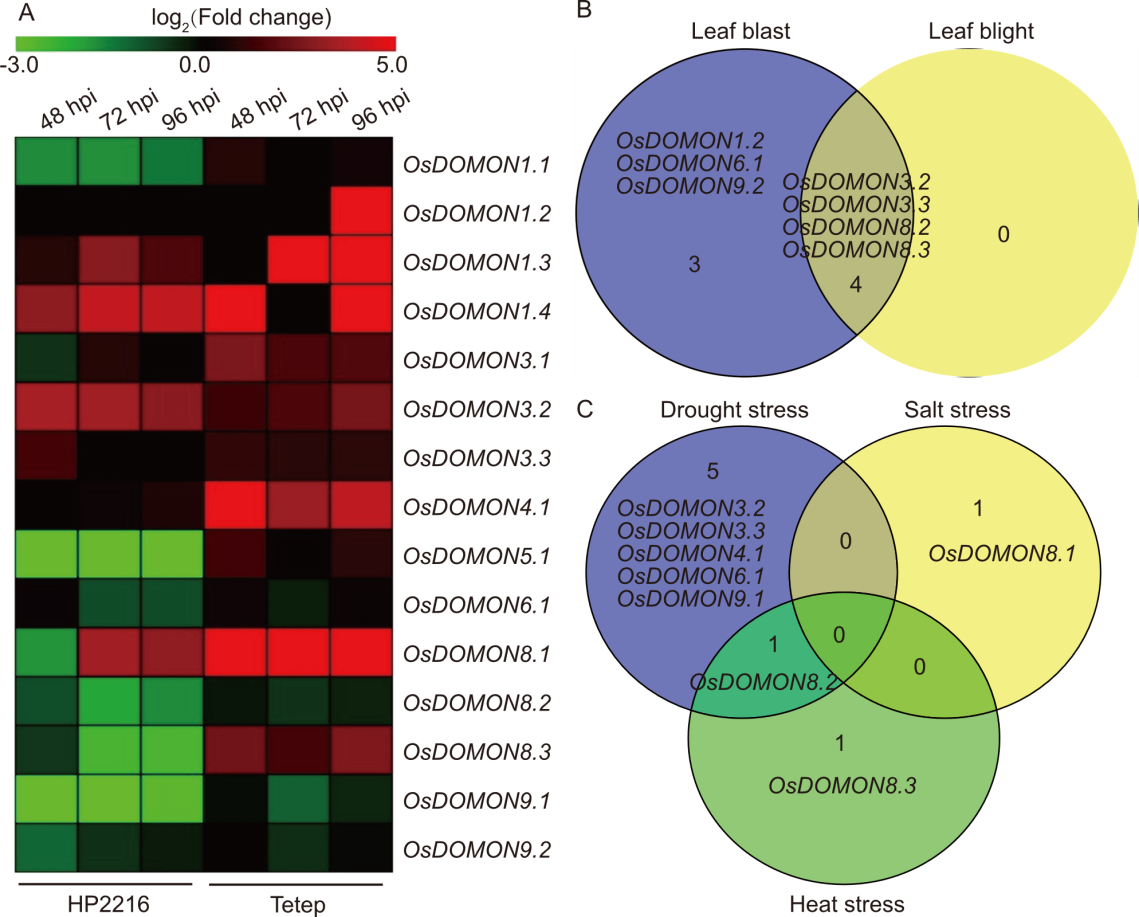
Fig. 4. Expression profiles of OsDOMON genes based on RNA-seq data in biotic and abiotic stresses. A, Heatmap displays the expression profiles of OsDOMON genes based on RNA-seq data of rice panicle tissues infected with Magnaporthe oryzae at 48, 72, and 96 h post-infection (hpi). The heatmap was constructed using TBtools. hpi, Hours post inoculation. B, Venn diagram representing the gene expression patterns of the OsDOMON genes in different biotic stresses using publicly available RNA-seq data from the RiceMetaSysB database. C, Gene expression patterns of the OsDOMON genes in different abiotic stresses using publicly available RNA-seq data from the RiceMetaSys and RiceMetaSysHRG databases.
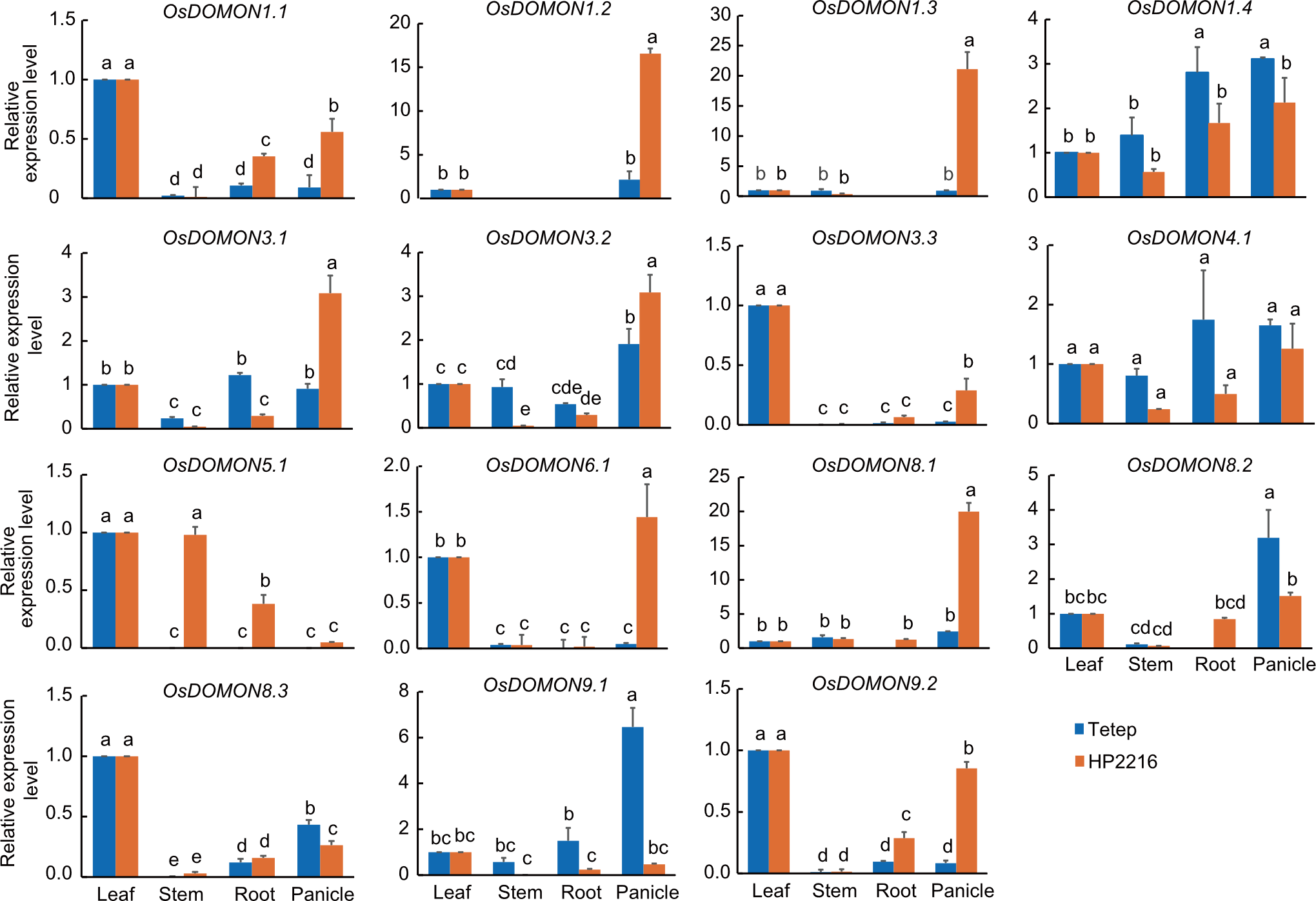
Fig. 5. Expression analysis of OsDOMON genes in different tissues (leaf, stem, root, and panicle) at the panicle stage of Tetep and HP2216 rice cultivars using qRT-PCR. Gene expression was analyzed in different tissues, with leaf tissue taken as the control for relative expression. Ubiquitin was used as the internal control. Data are mean ± SE (n = 3). The lowercase letters above bars indicate significant differences at the 0.05 level by the least significant difference test.
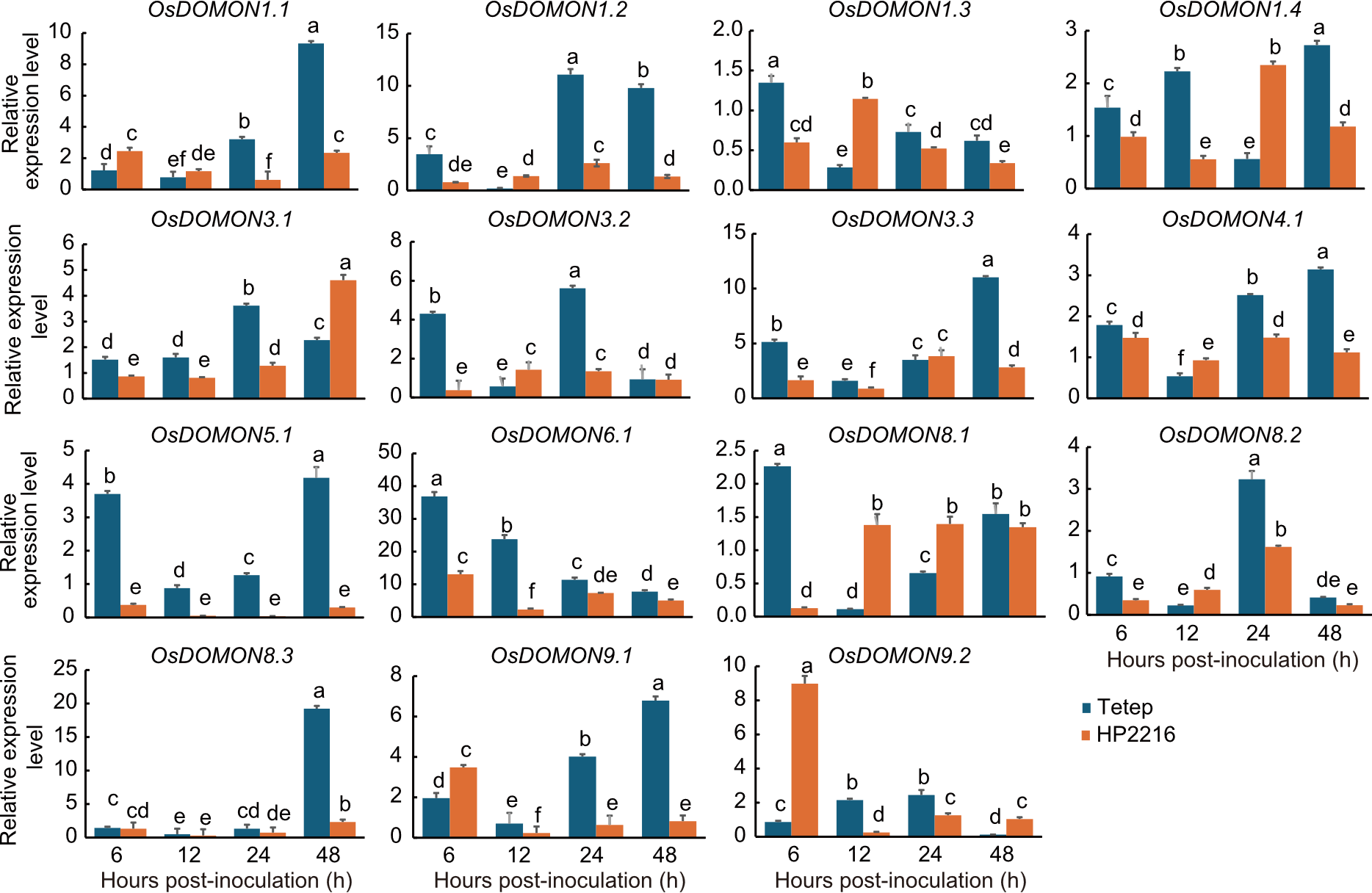
Fig. 6. Expression analysis of OsDOMON genes of Tetep and HP2216 after Magnaporthe oryzae infection at 6, 12, 24, and 48 hours post-inoculation intervals using qRT-PCR. Gene expression was analyzed in leaf tissues at the seedling stage, relative to 0 post-inoculation (at the time of infection). Ubiquitin was used as the internal control. Data are mean ± SE (n = 3). The lowercase letters above bars indicate significant differences at the 0.05 level by the least significant difference test.
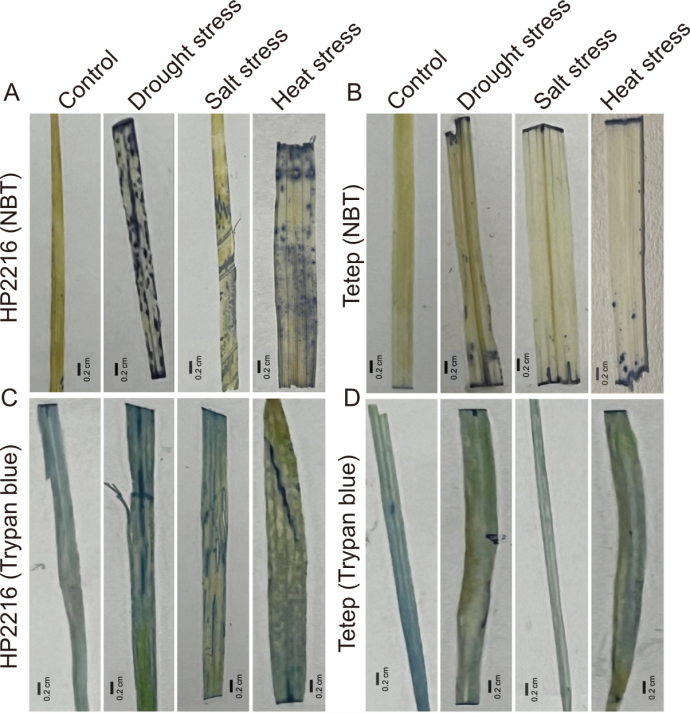
Fig. 7. Detection of reactive oxygen species accumulation by nitroblue tetrazolium (NBT A and B) staining and cell death by trypan blue staining (C and D) in rice leaves of Hp2216 (A and C) and Tetep (B and D) under abiotic stress conditions.
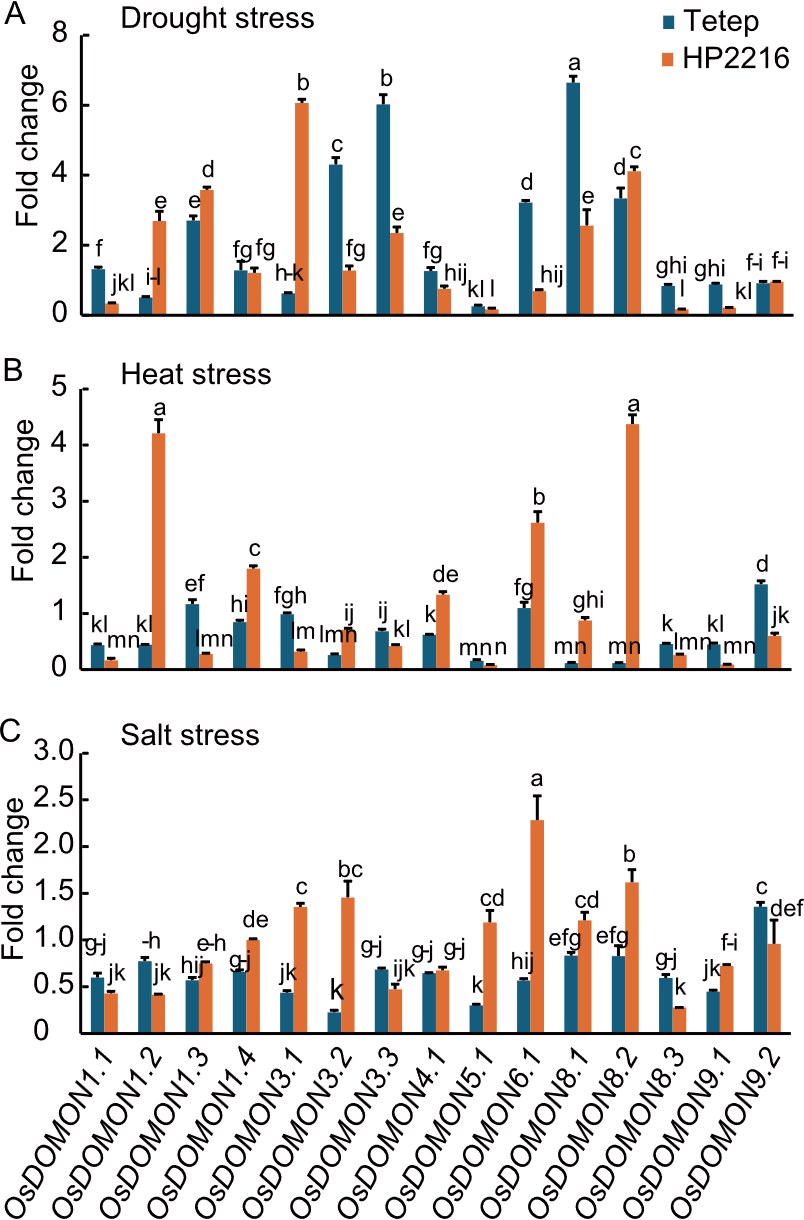
Fig. 8. Expression analysis of OsDOMON genes in Tetep and HP2216 under drought (A), heat (B), and salt (C) stresses using qRT-PCR. Gene expression was analyzed in leaf tissues, relative to unstressed leaf tissue. Ubiquitin was used as the internal control. Data are mean ± SE (n = 3). The lowercase letters above bars indicate significant differences at the 0.05 level by the least significant difference test.
| [1] | Aravind L. 2001. DOMON: An ancient extracellular domain in dopamine β-monooxygenase and other proteins. Trends Biochem Sci, 26(9): 524-526. |
| [2] | Asard H, Barbaro R, Trost P, et al. 2013. Cytochromes b561: Ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal, 19(9): 1026-1035. |
| [3] | Bailey T L, Boden M, Buske F A, et al. 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res, 37: W202-W208. |
| [4] | Baldoni E, Genga A, Cominelli E. 2015. Plant MYB transcription factors: Their role in drought response mechanisms. Int J Mol Sci, 16(7): 15811-15851. |
| [5] | Barrs H D, Weatherley P E. 1962. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci, 15(3): 413. |
| [6] | Biniek C, Heyno E, Kruk J, et al. 2017. Role of the NAD(P)H quinone oxidoreductase NQR and the cytochrome b AIR12 in controlling superoxide generation at the plasma membrane. Planta, 245(4): 807-817. |
| [7] | Brisson L F, Tenhaken R, Lamb C. 1994. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell, 6(12): 1703-1712. |
| [8] | Chaudhari S S, Moussian B, Specht C A, et al. 2014. Functional specialization among members of Knickkopf family of proteins in insect cuticle organization. PLoS Genet, 10(8): e1004537. |
| [9] | Chen C J, Chen H, Zhang Y, et al. 2020. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant, 13(8): 1194-1202. |
| [10] | Chen X Q, Wang F, Hyun J Y, et al. 2016. Recent progress in the development of fluorescent, luminescent and colorimetric probes for detection of reactive oxygen and nitrogen species. Chem Soc Rev, 45(10): 2976-3016. |
| [11] | Clúa J, Montpetit J, Jimenez-Sandoval P, et al. 2024. A CYBDOM protein impacts iron homeostasis and primary root growth under phosphate deficiency in Arabidopsis. Nat Commun 15: 423. |
| [12] | Costa A, Barbaro M R, Sicilia F, et al. 2015. AIR12, a b-type cytochrome of the plasma membrane of Arabidopsis thaliana is a negative regulator of resistance against Botrytis cinerea. Plant Sci, 233: 32-43. |
| [13] | Couch B C, Kohn L M. 2002. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia, 94(4): 683-693. |
| [14] | Deng P, Cao C J, Shi X Y, et al. 2023. OsCYBDOMG1, a cytochrome b561 domain-containing protein, regulates salt tolerance and grain yield in rice. Theor Appl Genet, 136(4): 76. |
| [15] | Devanna B N, Jain P, Solanke A U, et al. 2022. Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J Fungi, 8(6): 584. |
| [16] | Dong L Y, Liu S F, Kyaing M S, et al. 2020. Identification and fine mapping of Pi69(t), a new gene conferring broad-spectrum resistance against Magnaporthe oryzae from Oryza glaberrima steud. Front Plant Sci, 11: 1190. |
| [17] | Dong O X, Ronald P C. 2019. Genetic engineering for disease resistance in plants: Recent progress and future perspectives. Plant Physiol, 180(1): 26-38. |
| [18] | Edgar R C. 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5: 113. |
| [19] | Eragam A, Shukla V, Kola V S, et al. 2022. Yield-associated putative gene regulatory networks in Oryza sativa L. subsp. indica and their association with high-yielding genotypes. Mol Biol Rep, 49(8): 7649-7663. |
| [20] | Erpen L, Devi H S, Grosser J W, et al. 2018. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult, 132(1): 1-25. |
| [21] | FAO. 2024. Global forest resources assessment 2024:Main report. [2024-12-24]. https://www.fao.org/forest-resources-assessment/en/. |
| [22] | Fiyaz R A, Shivani D, Chaithanya K, et al. 2022. Genetic improvement of rice for bacterial blight resistance: Present status and future prospects. Rice Sci, 29(2): 118-132. |
| [23] | Gasteiger E, Hoogland C, Gattiker A, et al. 2005. Protein identification and analysis tools on the ExPASy server. In: Waller J M. The Proteomics Protocols Handbook. Totowa, NJ, USA: Humana Press: 571-607. |
| [24] | Gibson S W, Todd C D. 2015. Arabidopsis AIR12 influences root development. Physiol Mol Biol Plants, 21(4): 479-489. |
| [25] | Horton P, Park K J, Obayashi T, et al. 2007. WoLF PSORT: Protein localization predictor. Nucleic Acids Res, 35: W585-W587. |
| [26] | Hu B, Jin J P, Guo A Y, et al. 2015. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics, 31(8): 1296-1297. |
| [27] | Hückelhoven R. 2007. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol, 45: 101-127. |
| [28] | Iyer L M, Anantharaman V, Aravind L. 2007. The DOMON domains are involved in heme and sugar recognition. Bioinformatics, 23(20): 2660-2664. |
| [29] | Jagadish S V K, Craufurd P Q, Wheeler T R. 2007. High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot, 58(7): 1627-1635. |
| [30] | Jain P, Singh P K, Kapoor R, et al. 2017. Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene. Front Plant Sci, 8: 93. |
| [31] | Jain P, Dubey H, Singh P K, et al. 2019. Deciphering signalling network in broad spectrum Near Isogenic Lines of rice resistant to Magnaporthe oryzae. Sci Rep, 9(1): 16939. |
| [32] | Jayaraman K, Venkat Raman K, Sevanthi A M, et al. 2021. Stress-inducible expression of Chalcone isomerase2 gene improves accumulation of flavonoids and imparts enhanced abiotic stress tolerance to rice. Environ Exp Bot, 190: 104582. |
| [33] | Jiang M, Zhang J. 2003. Cross-talk between calcium and reactive oxygen species originated from NADPH oxidase in abscisic acid-induced antioxidant defence in leaves of maize seedlings. Plant Cell Environ, 26(6): 929-939. |
| [34] | Kariola T, Brader G, Li J, et al. 2005. Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell, 17(1): 282-294. |
| [35] | Karkute S G, Kumar V, Tasleem M, et al. 2022. Genome-wide analysis of von Willebrand factor A gene family in rice for its role in imparting biotic stress resistance with emphasis on rice blast disease. Rice Sci, 29(4): 375-384. |
| [36] | Karkute S G, Sevanthi A M, Solanke A U. 2023. Evolutionary relationship and structural analysis of blast resistance associated novel Osvwa36 and Osvwa37 genes in cultivated and wild species of rice. Int J Biol Resour Stress Manag, 14: 125-131. |
| [37] | Kloer D P, Hagel C, Heider J, et al. 2006. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure, 14(9): 1377-1388. |
| [38] | Krogh A, Larsson B, von Heijne G, et al. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol, 305(3): 567-580. |
| [39] | Krzywinski M, Schein J, Birol I, et al. 2009. Circos: An information aesthetic for comparative genomics. Genome Res, 19(9): 1639-1645. |
| [40] | Kumar A, Kumar V, Maheshwari C, et al. 2021. Nuclear localization and target genes analysis of an ABA and drought-responsive transcription factor (RDA1). Indian J Biotechnol, 20(4): 364-374. |
| [41] | Kumar A, Kumar S, Venkatesh K, et al. 2022. Physio-molecular traits of contrasting bread wheat genotypes associated with 15N influx exhibiting homeolog expression bias in nitrate transporter genes under different external nitrate concentrations. Planta, 255(5): 104. |
| [42] | Kumar A, Maheshwari C, Phogat S, et al. 2024. Impact of biotic and abiotic interaction on crop plants. In: Climate-Resilient Agriculture. New York, USA: Apple Academic Press: 107-134. |
| [43] | Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 35(6): 1547-1549. |
| [44] | Kumar V, Jain P, Venkadesan S, et al. 2021. Understanding rice- Magnaporthe oryzae interaction in resistant and susceptible cultivars of rice under panicle blast infection using a time-course transcriptome analysis. Genes, 12(2): 301. |
| [45] | Kumar V, Kumar A, Tewari K, et al. 2022b. Isolation and characterization of drought and ABA responsive promoter of a transcription factor encoding gene from rice. Physiol Mol Biol Plants, 28(10): 1813-1831. |
| [46] | Kumar V, Singh P K, Karkute S G, et al. 2022a. Identification of novel resources for panicle blast resistance from wild rice accessions and mutants of cv. Nagina 22 by syringe inoculation under field conditions. 3 Biotech, 12(2): 53. |
| [47] | Kumari P, Devi L L, Kumar A, et al. 2022. Differential response of rice genotypes to nitrogen availability is associated with the altered nitrogen metabolism and ionomic balance. Environ Exp Bot, 198: 104847. |
| [48] | Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res, 49(W1): W293-W296. |
| [49] | Letunic I, Khedkar S, Bork P. 2021. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res, 49: D458-D460. |
| [50] | Li Z, Peng M X, Power D M, et al. 2021. RNAi-mediated knock-down of the dopamine beta-hydroxylase gene changes growth of razor clams. Comp Biochem Physiol Part B Biochem Mol Biol, 252: 110534. |
| [51] | Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402-408. |
| [52] | Madeira F, Madhusoodanan N, Lee J, et al. 2024. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res, 52: W521-W525. |
| [53] | Mahalingam R, Jambunathan N, Gunjan S K, et al. 2006. Analysis of oxidative signalling induced by ozone in Arabidopsis thaliana. Plant Cell Environ, 29(7): 1357-1371. |
| [54] | Meng E C, Goddard T D, Pettersen E F, et al. 2023. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci, 32(11): e4792. |
| [55] | Mistry J, Chuguransky S, Williams L, et al. 2021. Pfam:The protein families database in 2021. Nucleic Acids Res, 49: D412-D419. |
| [56] | Paysan-Lafosse T, Blum M, Chuguransky S, et al. 2023. InterPro in 2022. Nucleic Acids Res, 51: D418-D427. |
| [57] | Ponting C P, Mott R, Bork P, et al. 2001. Novel protein domains and repeats in Drosophila melanogaster: Insights into structure, function, and evolution. Genome Res, 11(12): 1996-2008. |
| [58] | Preger V, Tango N, Marchand C, et al. 2009. Auxin-responsive genes AIR12 code for a new family of plasma membrane b-type cytochromes specific to flowering plants. Plant Physiol, 150(2): 606-620. |
| [59] | Qiu D Y, Xiao J, Ding X H, et al. 2007. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact, 20(5): 492-499. |
| [60] | Qiu Y P, Jing S J, Fu J, et al. 2004. Cloning and analysis of expression profile of 13 WRKY genes in rice. Chin Sci Bull, 49: 2159-2168. |
| [61] | Rehman S, Mahmood T. 2015. Functional role of DREB and ERF transcription factors: Regulating stress-responsive network in plants. Acta Physiol Plant, 37(9): 178. |
| [62] | Sandhu M, Sureshkumar V, Prakash C, et al. 2017. RiceMetaSys for salt and drought stress responsive genes in rice: A web interface for crop improvement. BMC Bioinformatics, 18(1): 432. |
| [63] | Sharma T R, Rai A K, Gupta S K, et al. 2012. Rice blast management through host-plant resistance: Retrospect and prospects. Agric Res, 1(1): 37-52. |
| [64] | Singh B K, Venkadesan S, Ramkumar M K, et al. 2023. Meta-analysis of microarray data and their utility in dissecting the mapped QTLs for heat acclimation in rice. Plants, 12(8): 1697. |
| [65] | Sinha S K, Kumar A, Tyagi A, et al. 2020. Root architecture traits variation and nitrate-influx responses in diverse wheat genotypes under different external nitrogen concentrations. Plant Physiol Biochem, 148: 246-259. |
| [66] | Soltész A, Vágújfalvi A, Rizza F, et al. 2012. The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. J Appl Genet, 53(2): 133-143. |
| [67] | Sonah H, Deshmukh R K, Chand S, et al. 2012. Molecular mapping of quantitative trait loci for flag leaf length and other agronomic traits in rice (Oryza sativa). Cereal Res Commun, 40(3): 362-372. |
| [68] | Sureshkumar V, Dutta B, Kumar V, et al. 2019. RiceMetaSysB: A database of blast and bacterial blight responsive genes in rice and its utilization in identifying key blast-resistant WRKY genes. Database, 2019: baz015. |
| [69] | Tarun J A, Mauleon R, Arbelaez J D, et al. 2020. Comparative transcriptomics and co-expression networks reveal tissue- and genotype-specific responses of qDTYs to reproductive-stage drought stress in rice (Oryza sativa L.). Genes, 11(10): 1124. |
| [70] | Tian F, Yang D C, Meng Y Q, et al. 2020. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res, 48: D1104-D1113. |
| [71] | Tsubaki M, Takeuchi F, Nakanishi N. 2005. Cytochrome b561 protein family: Expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta, 1753(2): 174-190. |
| [72] | Varadi M, Bertoni D, Magana P, et al. 2024. AlphaFold protein structure database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res, 52: D368-D375. |
| [73] | Voorrips R E. 2002. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered, 93(1): 77-78. |
| [74] | Wang Q, Shi H F, Huang R S, et al. 2021. AIR12 confers cold tolerance through regulation of the CBF cold response pathway and ascorbate homeostasis. Plant Cell Environ, 44(5): 1522-1533. |
| [75] | Yan J W, Liu Y, Yang L, et al. 2021. Cell wall β-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol Plant, 14(3): 411-425. |
| [76] | Yan X, Tang B Z, Ryder L S, et al. 2023. The transcriptional landscape of plant infection by the rice blast fungus Magnaporthe oryzae reveals distinct families of temporally co-regulated and structurally conserved effectors. Plant Cell, 35(5): 1360-1385. |
| [77] | Yin Z, Chen J, Zeng L, et al. 2000. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact, 13(8): 869-876. |
| [78] | Yoshida S, Coronel V. 1976. Nitrogen nutrition, leaf resistance, and leaf photosynthetic rate of the rice plant. Soil Sci Plant Nutr, 22(2): 207-211. |
| [79] | Yu C S, Lin C J, Hwang J K. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci, 13(5): 1402-1406. |
| [80] | Yu R R, Zhang R, Liu W M, et al. 2022. The DOMON domain protein LmKnk contributes to correct chitin content, pore canal formation and lipid deposition in the cuticle of Locusta migratoria during moulting. Insect Mol Biol, 31(2): 127-138. |
| [81] | Zhang C H, Shangguan L F, Ma R J, et al. 2012. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet Mol Res, 11(4): 4789-4809. |
| [82] | Zhang X, Long Y, Huang J J, et al. 2020. OsNAC45 is involved in ABA response and salt tolerance in rice. Rice, 13(1): 79. |
| [1] | Daisy Wilson, Valeria Gonzalez, Hamidreza Sharifan. Evaluating Efficacy of ZnO and MgO Nanoparticles on Post-Harvested Rice to Enhance Food Security Against Agroterrorism [J]. Rice Science, 2025, 32(5): 717-726. |
| [2] | Li Haifeng, Fan Jiayi. Functions of Rice E3 Ubiquitin Ligases in Response to Environmental Stress and in Regulating Grain Size [J]. Rice Science, 2025, 32(5): 649-657. |
| [3] | Wu Zhaozhong, Zhong Zhengzheng, Xu Peng, Liu Ling, Wang Beifang, Yang Qinqin, Wen Xiaoxia, Ma Guifang, Luo Mili, Zhang Yingxin, Liu Qun’en, Peng Zequn, Zhan Xiaodeng, Cao Liyong, Cheng Shihua, Wu Weixun. OsELF3.1-OsCATA-Ghd7 Pathway Regulates Rice Heading [J]. Rice Science, 2025, 32(5): 658-672. |
| [4] | Pan Pan, Guo Wenlong, Li Hengbo, Shao Yifan, Guo Zhihao, Jin Ye, Cheng Yanrong, Yu Guoping, Fu Zhenshi, Hu Lin, Zheng Xiaoming, Zhou Guomin, Zhang Jianhua. Accelerating Wild Rice Disease-Resistant Germplasm Exploration: Artificial Intelligence (AI)-Powered Wild Rice Blast Disease Level Evaluation and Disease-Resistance Identification [J]. Rice Science, 2025, 32(5): 727-746. |
| [5] | Sabarinathan Selvaraj, Parameswaran Chidambaranathan, Goutam Kumar Dash, Priyadarsini Sanghamitra, Kishor Pundlik Jeughale, Cayalvizhi Balasubramaniasai, Devraj Lenka, Basavantraya Navadagi Devanna, Seenichamy Rathinam Prabhukarthikeyan, Sanghamitra Samantaray, Amaresh Kumar Nayak. Long-Range Admixture Linkage Disequilibrium and Allelic Responses of Sub1 and TPP7 under Consecutive Stress in Rice Validated Through Mendelian Randomization [J]. Rice Science, 2025, 32(5): 704-716. |
| [6] | Yong Jin Choi, Sun-Hwa Ha. Metabolic Engineering in Rice for Functional Metabolite Production [J]. Rice Science, 2025, 32(4): 475-498. |
| [7] | Dinuka Nuwan Tharaka, Nadeeka D. Tissera, Gayan Priyadarshana, Damayanthi Dahanayake. A Comprehensive Review of Hierarchical Porous Carbon Synthesis from Rice Husk [J]. Rice Science, 2025, 32(4): 499-511. |
| [8] | Li Xinyan, Weng Lüshui, Xiao Youlun, Li Jinjiang, Deng Lihua, Liu Qing, Kang Weiwei, Duan Yaping, Yang Daji, Xiao Guoying. Characteristic Analysis of Penta-Resistance Restorer Line for Hybrid Rice [J]. Rice Science, 2025, 32(4): 512-524. |
| [9] | Zhou Lin, Jiang Hong, Huang Long, Li Ziang, Yao Zhonghao, Li Linhan, Ji Kangwei, Li Yijie, Tang Haijuan, Cheng Jinping, Bao Yongmei, Huang Ji, Zhang Hongsheng, Chen Sunlu. Genome-Wide Association Study of Brown Rice Weight Identifies an RNA-Binding Protein Antagonistically Regulating Grain Weight and Panicle Number [J]. Rice Science, 2025, 32(4): 525-536. |
| [10] | Ratan Kumar Ganapati, Chen Kai, Zhao Xiuqin, Zheng Tianqing, Zhang Fan, Zhai Laiyuan, Xu Jianlong. Genome-Wide Association Study and Haplotype Analysis Jointly Identify New Candidate Genes for Alkaline Tolerance at Seedling Stage in Rice [J]. Rice Science, 2025, 32(4): 537-548. |
| [11] | Hou Yuxuan, Zhu Jie, Lu Chenglong, Fan Libo, Liang Mengqi, Zhang Xiaobo, Cheng Benyi, Xu Xia, Gong Junyi. A Recombinase-Aided Amplification-Lateral Flow Dipstick Detection Technique for Early On-Site Diagnosis of Bacterial Blight Caused by Xanthomonas oryzae pv. oryzae in Rice [J]. Rice Science, 2025, 32(4): 575-584. |
| [12] | Chen Su, Ma Feilong, Chen Jiaoyang, Qi Man, Wei Qianshu, Tao Zhihuan, Sun Bo. Function of R2R3-Type Myeloblastosis Transcription Factors in Plants [J]. Rice Science, 2025, 32(3): 307-321. |
| [13] | Yang Yajun, Lu Yanhui, Tian Junce, Zheng Xusong, Guo Jiawen, Liu Xiaowei, Lü Zhongxian, Xu Hongxing. Sustainable Management Strategies for Rice Leaffolder, Cnaphalocrocis medinalis (Guenée): Progress and Prospects [J]. Rice Science, 2025, 32(3): 322-338. |
| [14] | Xie Yuhao, Xie Wenya, Zhao Jianhua, Xue Xiang, Cao Wenlei, Shi Xiaopin, Wang Zhou, Wang Yiwen, Wang Guangda, Feng Zhiming, Hu Keming, Chen Xijun, Chen Zongxiang, Zuo Shimin. OsERF7 Negatively Regulates Resistance to Sheath Blight Disease by Inhibiting Phytoalexin Biosynthesis [J]. Rice Science, 2025, 32(3): 367-379. |
| [15] | Chaemyeong Lim, Sae Hyun Lee, Haeun Lee, So-Yon Park, Kiyoon Kang, Hyeryung Yoon, Tae-Jin Yang, Gary Stacey, Nam-Chon Paek, Sung-Hwan Cho. Global Transcriptome Analysis of Rice Seedlings in Response to Extracellular ATP [J]. Rice Science, 2025, 32(3): 380-399. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||