
Rice Science ›› 2020, Vol. 27 ›› Issue (1): 10-20.DOI: 10.1016/j.rsci.2019.12.002
• Research Paper • Previous Articles Next Articles
Xixu Peng1,2,3( ), Ting Xiao1, Jiao Meng1, Zong Tao1, Dinggang Zhou1,2, Xinke Tang1,3, Haihua Wang1,2,3(
), Ting Xiao1, Jiao Meng1, Zong Tao1, Dinggang Zhou1,2, Xinke Tang1,3, Haihua Wang1,2,3( )
)
Received:2018-12-27
Accepted:2019-05-09
Online:2020-01-28
Published:2019-09-30
Xixu Peng, Ting Xiao, Jiao Meng, Zong Tao, Dinggang Zhou, Xinke Tang, Haihua Wang. Differential Expression of Rice Valine-Qlutamine Gene Family in Response to Nitric Oxide and Regulatory Circuit of OsVQ7 and OsWRKY24[J]. Rice Science, 2020, 27(1): 10-20.
Add to citation manager EndNote|Ris|BibTeX
| Gene | RAP_ID | Chromosome | No. of amino acids (aa) | Exon number | Fold change of gene expression a | ||
| T1/T0 | T6/T0 | T12/T0 | |||||
| OsVQ1 | Os01g0278000 | 1 | 235 | 1 | 1.10 | 3.30 | 8.03 |
| OsVQ4 | Os01g0808900 | 1 | 184 | 1 | 0.37 | 0.81 | 0.92 |
| OsVQ5 | Os02g0173200 | 2 | 229 | 1 | 2.07 | 1.14 | 1.64 |
| OsVQ7 | Os02g0251900 | 2 | 126 | 1 | 8.75 | 1.75 | 2.84 |
| OsVQ9 | Os02g0753700 | 2 | 190 | 1 | 1.75 | 0.48 | 0.89 |
| OsVQ13 | Os03g0676400 | 3 | 143 | 1 | 5.47 | 1.11 | 0.56 |
| OsVQ16 | Os04g0645200 | 4 | 202 | 1 | 2.29 | 1.05 | 0.69 |
| OsVQ19 | Os05g0390800 | 5 | 169 | 2 | 0.47 | 0.65 | 1.07 |
| OsVQ22 | Os06g0530600 | 6 | 144 | 1 | 4.08 | 1.00 | 1.52 |
| OsVQ24 | Os06g0618700 | 6 | 188 | 1 | 0.99 | 0.55 | 0.27 |
| OsVQ30 | Os07g0686600 | 7 | 218 | 1 | 2.29 | 10.57 | 2.50 |
| OsVQ31 | Os07g0687400 | 7 | 130 | 2 | 1.73 | 3.92 | 1.73 |
| OsVQ33 | Os08g0409500 | 8 | 426 | 1 | 3.12 | 1.70 | 1.92 |
| OsVQ40 | Os03g0189900 | 3 | 248 | 1 | 1.22 | 1.86 | 3.50 |
Table 1 Differentially expressed rice VQ genes under nitric oxide treatment.
| Gene | RAP_ID | Chromosome | No. of amino acids (aa) | Exon number | Fold change of gene expression a | ||
| T1/T0 | T6/T0 | T12/T0 | |||||
| OsVQ1 | Os01g0278000 | 1 | 235 | 1 | 1.10 | 3.30 | 8.03 |
| OsVQ4 | Os01g0808900 | 1 | 184 | 1 | 0.37 | 0.81 | 0.92 |
| OsVQ5 | Os02g0173200 | 2 | 229 | 1 | 2.07 | 1.14 | 1.64 |
| OsVQ7 | Os02g0251900 | 2 | 126 | 1 | 8.75 | 1.75 | 2.84 |
| OsVQ9 | Os02g0753700 | 2 | 190 | 1 | 1.75 | 0.48 | 0.89 |
| OsVQ13 | Os03g0676400 | 3 | 143 | 1 | 5.47 | 1.11 | 0.56 |
| OsVQ16 | Os04g0645200 | 4 | 202 | 1 | 2.29 | 1.05 | 0.69 |
| OsVQ19 | Os05g0390800 | 5 | 169 | 2 | 0.47 | 0.65 | 1.07 |
| OsVQ22 | Os06g0530600 | 6 | 144 | 1 | 4.08 | 1.00 | 1.52 |
| OsVQ24 | Os06g0618700 | 6 | 188 | 1 | 0.99 | 0.55 | 0.27 |
| OsVQ30 | Os07g0686600 | 7 | 218 | 1 | 2.29 | 10.57 | 2.50 |
| OsVQ31 | Os07g0687400 | 7 | 130 | 2 | 1.73 | 3.92 | 1.73 |
| OsVQ33 | Os08g0409500 | 8 | 426 | 1 | 3.12 | 1.70 | 1.92 |
| OsVQ40 | Os03g0189900 | 3 | 248 | 1 | 1.22 | 1.86 | 3.50 |
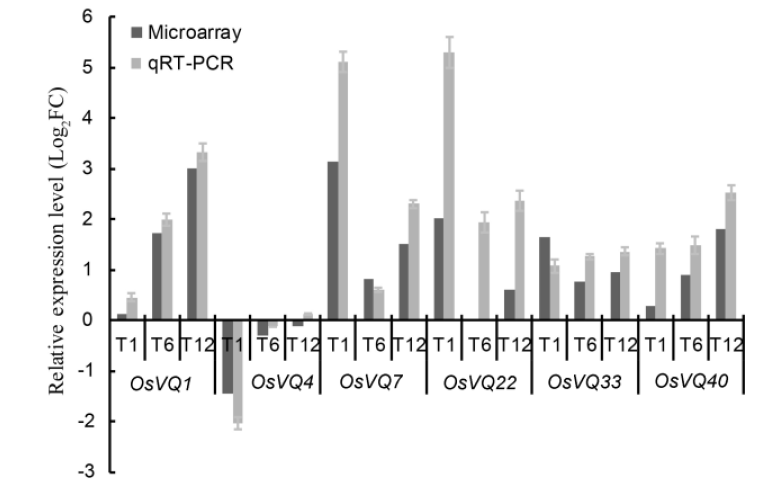
Fig. 1. Quantitative real-time PCR (qRT-PCR) verification of VQ gene expression in microarray assay. ===Log2FC, Log2 transformed fold change. T1, T6 and T12 refer to rice samples harvested at 1, 6 and 12 h after nitric oxide treatment, respectively.===Data represent as Mean ± SE (n = 3).
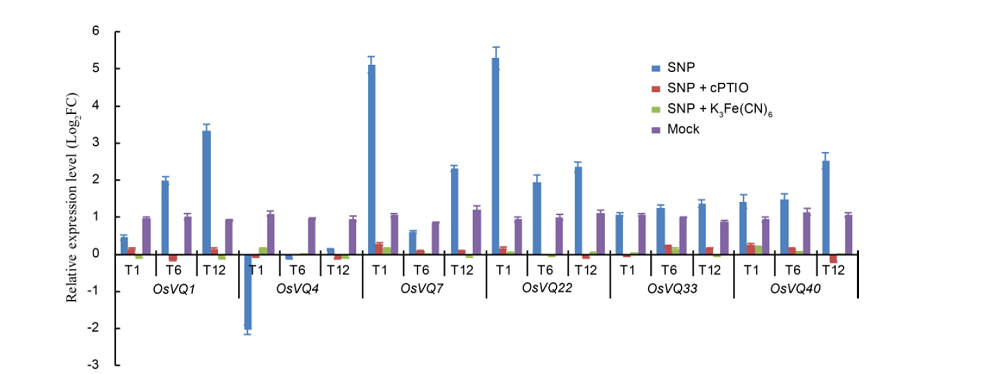
Fig. 2. Quantitative real-time PCR analysis for VQ gene expression of rice seedlings treated with cPTIO or K3Fe(CN)6. ===Log2FC, Log2 transformed fold change. T1, T6 and T12 refer to rice samples harvested at 1, 6 and 12 h after nitric oxide treatment, respectively. ===0.1% Tween-80 was used as mock. Data represent as Mean ± SE (n = 3).
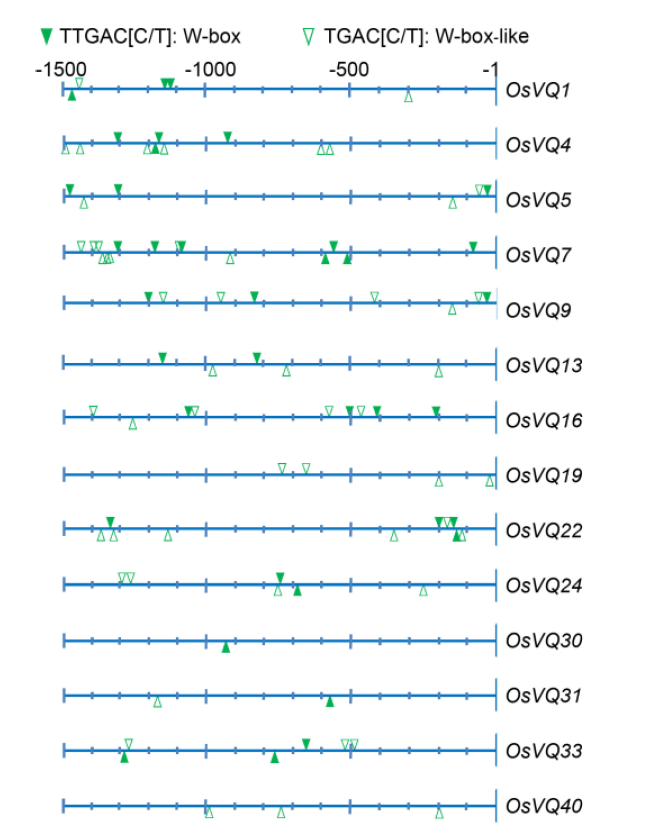
Fig. 3. Illustration of W-box or W-box-like cis-elments in the promoter regions of nitric oxide-responsive rice VQ genes.===For each gene, the promoter region of 1 500 bp upstream from the transcription start site (arrow) is shown.
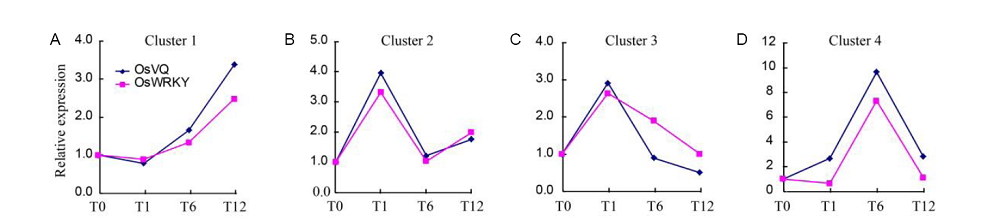
Fig. 4. Cluster analysis of expression patterns of nitric oxide-responsive rice VQ and WRKY genes.===A, Cluster 1 including OsVQ1, OsVQ4, OsVQ19, OsVQ40 and OsWRKY3, OsWRKY29, OsWRKY39, OsWRKY62, OsWRKY77, OsWRKY79, OsWRKY88, OsWRKY109. B, Cluster 2 including OsVQ5, OsVQ7, OsVQ9, OsVQ22, OsVQ33 and OsWRKY1, OsWRKY15, OsWRKY23, OsWRKY24, OsWRKY28, OsWRKY53, OsWRKY56, OsWRKY69, OsWRKY71, OsWRKY72, OsWRKY104. C, Cluster 3 including OsVQ13, OsVQ16, OsVQ24 and OsWRKY8, OsWRKY12, OsWRKY13, OsWRKY21, OsWRKY42, OsWRKY60, OsWRKY67. D, Cluster 4 including OsVQ30, OsVQ31 and OsWRKY50. ===Genes were divided into four clusters based on their expression levels using the hierachical clustering (HCL) algorithm.===T0, T1, T6 and T12 refer to rice samples harvested at 0, 1, 6 and 12 h after nitric oxide treatment, respectively.
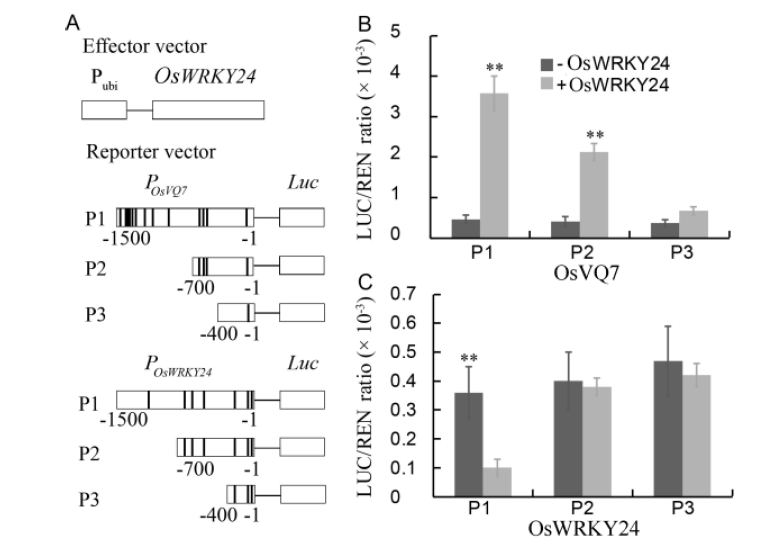
Fig. 5. Transient assay for the interaction between OsWRKY24 and promoters of OsVQ7 and OsWRKY24 itself in Nicotiana benthamiana.===A, Diagram of the promoters of OsVQ7 and OsWRKY24, showing W-box or W-box-like elements (bold vertical lines) in different regions. B and C, Transient expression assay of OsVQ7 and OsWRKY24 in tobacco leaves. ===Luc, Firefly luciferase activity; REN, Renilla luciferase activity (used as the control). P1, Full promoter of OSWRKY24 or OsVQ7 (-1 to -1500); P2, Truncated promoter of OSWRKY24 or OsVQ7 (-1 to -700); P3, Truncated promoter of OSWRKY24 or OsVQ7 (-1 to -400).===Data represent as Mean ± SE (n = 3). *, P < 0.05. ** P < 0.01.
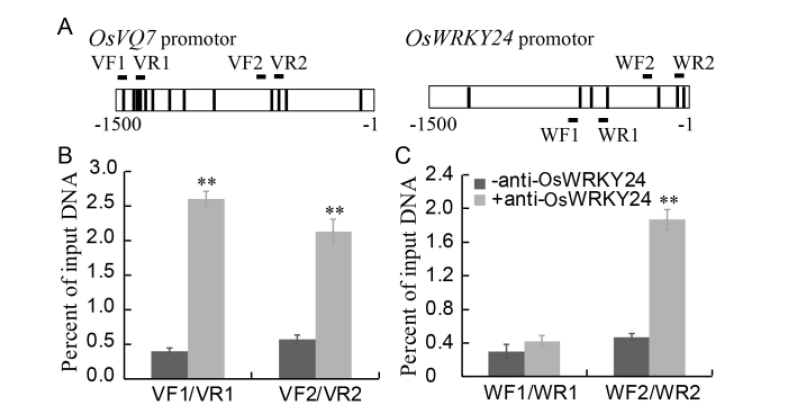
Fig. 6. ChIP assay for the binding of OsWRKY24 to the promoters of OsVQ7 and OsWRKY24 itself in rice.===A, Diagram of OsVQ7 and OsWRKY24 promoters showing W-box or W-box-like elements (bold vertical lines) in different regions. B, Binding of OsWRKY24 to OsVQ7 promoter. C, Binding of OsWRKY24 to its own gene promoter. ===Data represent as Mean ± SE (n = 3). **, P < 0.01. ===The extracts from the leaves of rice seedlings (three-leaf-stage) were used for ChIP assay
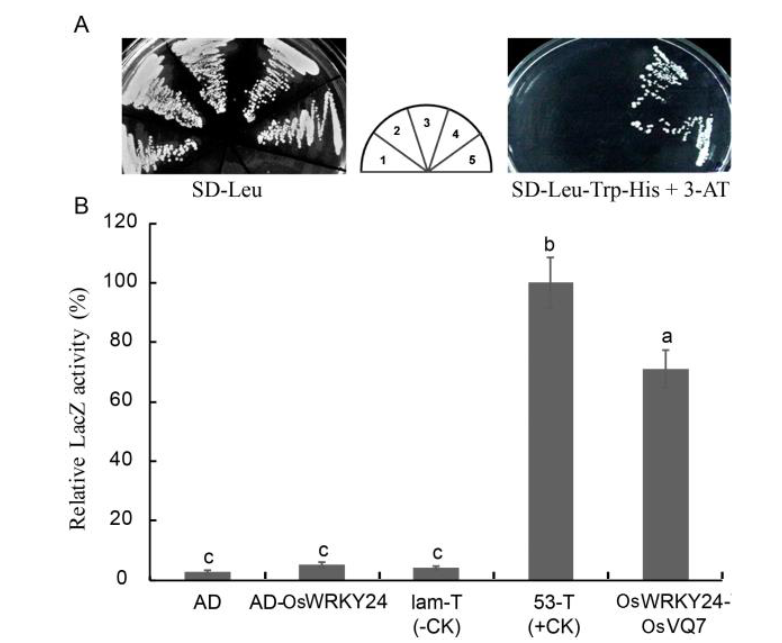
Fig. 7. Interaction of OsVQ7 with OsWRKY24 in yeast cells.===A, Left panel, yeast cells carrying different constructs were grown for 2 d at 30 ºC in SD-Leu agar medium. Middle, schematic distribution of yeast cells carrying different vectors; 1, AD (empty vector pAD, negative control); 2, AD-OsWRKY24; 3, lam protein-T protein (negative control); 4, 53-T protein (positive control); 5, OsWRKY24-OsVQ7. Right panel, positive interaction between OsVQ7 and OsWRKY24 was verified in SD-Leu-Trp-His agar medium with 30 mmol/L 3-amino- 1,2,4-triazole (3-AT). B, Assay for β-galactosidase activity. The enzymatic activity of cells cotransformed with pBD-53 and pAD-T was set as 100%. ===Data represent as Mean ± SE (n = 3), and different lowercase letters above columns indicate significant differences based on the Duncan’s multiple range test (P < 0.05).
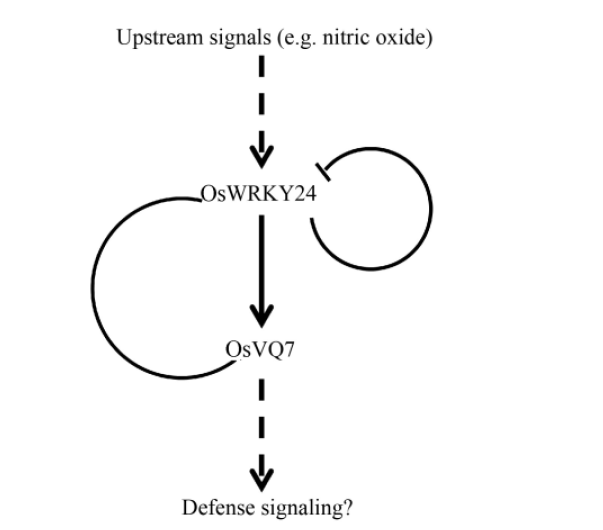
Fig. 8. Model of transcriptional regulatory circuit of rice OsWRKY24 and OsVQ7.===Solid arrow and T lines indicate induction and suppression of gene expression, respectively. The round line between OsWRKY24 and OsVQ7 indicates their interaction. Dotted arrows indicate predicted actions.
| [1] | Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen N H T, Zhu S J, Qiu J L, Micheelsen P, Rocher A, Petersen M, Newman M A, Nielsen H B, Hirt H, Somssich I, Mattsson O, Mundy J.2005. The MAP kinase substrate MKS1 is a regulator of plant defense responses.EMBO J, 24: 2579-2589. |
| [2] | Benhamed M, Bertrand C, Servet C, Zhou D X.2006. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell, 18(11): 2893-2903. |
| [3] | Chen L G, Song Y, Li S J, Zhang L P, Zou C S, Yu D Q.2012. The role of WRKY transcription factors in plant abiotic stresses.Biochim Biophys Acta, 1819(2): 120-128. |
| [4] | Cheng H T, Liu H N, Deng Y, Xiao J H, Li X H, Wang S P.2015. The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen.Plant Physiol, 167(3): 1087-1099. |
| [5] | Cheng Y, Zhou Y, Yang Y, Chi Y J, Zhou J Y, Chen J Y, Wang F, Fan B, Shi K, Zhou Y H, Yu J Q, Chen Z.2012. Structural and functional analysis of VQ motif-containing proteins inArabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol, 159(2): 810-825. |
| [6] | Chi Y J, Yang Y, Zhou Y, Zhou J, Fan B F, Yu J Q, Chen Z X.2013. Protein-protein interactions in the regulation of WRKY transcription factors.Mol Plant, 6(2): 287-300. |
| [7] | Dong J X, Chen C H, Chen Z X.2003. Expression profiles of theArabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol, 51(1): 21-37. |
| [8] | Dong Q L, Zhao S, Duan D Y, Tian Y, Wang Y P, Mao K, Zhou Z S, Ma F W.2018. Structural and functional analyses of genes encoding VQ proteins in apple.Plant Sci, 272: 208-219. |
| [9] | Eulgem T, Rushton P J, Robatzek S, Somssich I E.2000. The WRKY superfamily of plant transcription factors.Trends Plant Sci, 5(5): 199-206. |
| [10] | Eulgem T, Somssich I E.2007. Networks of WRKY transcription factors in defense signaling.Curr Opin Plant Biol, 10(4): 366-371. |
| [11] | Glazebrook J.2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens.Annu Rev Phytopathol, 43: 205-227. |
| [12] | Hellens R P, Allan A C, Friel E N, Bolitho K, Grafton K, Templeton M D, Karunairetnam S, Gleave A P, Laing W A.2005. Transient plant expression vectors for functional genomics, quantification of promoter activity and RNA silencing.Plant Methods, 1: 13. |
| [13] | Higo K, Ugawa Y, Iwamoto M, Korenaga T.1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999.Nucl Acids Res, 27(1): 297-300. |
| [14] | Hu Y R, Chen L G, Wang H P, Zhang L P, Wang F, Yu D Q.2013. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J, 74(5): 730-745. |
| [15] | Jiang S Y, Sevugan M, Ramachandran S.2018. Valine-glutamine (VQ) motif coding genes are ancient and non-plant-specific with comprehensive expression regulation by various biotic and abiotic stresses.BMC Genom, 19: 342. |
| [16] | Jing Y J, Lin R C.2015. The VQ motif-containing protein family of plant-specific transcriptional regulators.Plant Physiol, 169(1): 371-378. |
| [17] | Kim D Y, Kwon S I, Choi C, Lee H, Ahn I, Park S R, Bae S C, Lee S C, Hwang D J.2013. Expression analysis of rice VQ genes in response to biotic and abiotic stresses.Gene, 529(2): 208-214. |
| [18] | Lai Z B, Li Y, Wang F, Cheng Y, Fan B F, Yu J Q, Chen Z X.2011. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell, 23(10): 3824-3841. |
| [19] | Li N, Li X H, Xiao J H, Wang S P.2014. Comprehensive analysis of VQ motif-containing gene expression in rice defense responses to three pathogens.Plant Cell Rep, 33(9): 1493-1505. |
| [20] | Li Y L, Jing Y J, Li J J, Xu G, Lin R C.2014. Arabidopsis VQ MOTIFCONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME- INTERACTING FACTOR1. Plant Physiol, 164(4): 2068-2080. |
| [21] | Lin Y Y, Yu D Q.2012. Expression profiles ofArabidopsis thaliana VQ gene family in defense-related hormones treatments. Plant Divers Resour, 34(5): 509-518. (in Chinese with English abstract) |
| [22] | Meng J, Wang H H, Xiang J H, Jiang D, Peng X X, He H H.2016. Expression profiles of rice WRKY transcription factor gene family responsive to exogenous nitric oxide application.Chin J Rice Sci, 30(2): 111-120. (in Chinese with English abstract) |
| [23] | Palmieri M C, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C.2008. Nitric oxide-responsive genes and promoters inArabidopsis thaliana: A bioinformatics approach. J Exp Bot, 59(2): 177-186. |
| [24] | Pandey S P, Somssich I E.2009. The role of WRKY transcription factors in plant immunity.Plant Physiol, 150(4): 1648-1655. |
| [25] | Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman D W, Goldman S L.2004. Microarray analysis of nitric oxide responsive transcripts inArabidopsis. Plant Biotechnol J, 2(4): 359-366. |
| [26] | Pecher P, Eschen-Lippold L, Herklotz S, Kuhle K, Naumann K, Bethke G, Uhrig J, Weyhe M, Scheel D, Lee J.2014. TheArabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol, 203(2): 592-606. |
| [27] | Peng X X, Hu Y J, Tang X K, Zhou P L, Deng X B, Wang H H, Guo Z J.2012. Constitutive expression of riceWRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236(5): 1485-1498. |
| [28] | Peng X X, Wang H H, Jang J C, Xiao T, He H H, Jiang D, Tang X K.2016. OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance againstRhizoctonia solani. Rice, 9: 63. |
| [29] | Peng X X, Bai N N, Wang H H.2018. Isolation and expression profiles of cadmium stress-responsive rice WRKY15 transcription factor gene.Chin J Rice Sci, 32(2): 103-110. (in Chinese with English abstract) |
| [30] | Perruc E, Charpenteau M, Ramirez B C, Jauneau A, Galaud J P, Ranjeva R, Ranty B.2004. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance inArabidopsis thaliana seedlings. Plant J, 38(3): 410-420. |
| [31] | Petersen K, Qiu J L, Lütje J, Fiil B K, Hansen S, Mundy J, Petersen M.2010. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS One, 5(12): e14364. |
| [32] | Rokach L, Maimon O. Clustering methods.2005. Data Mining and Knowledge Discovery Handbook. Boston, MA, US: Springer: 321-352. |
| [33] | Ryu H S, Han M, Lee S K, Cho J I, Ryoo N, Heu S, Lee Y H, Bhoo S H, Wang G L, Hahn T R, Jeon J S.2006. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response.Plant Cell Rep, 25(8): 836-847. |
| [34] | Shannon P, Markiel A, Ozier O, Baliga N S, Wang J T, Ramage D, Amin N, Schwikowski B, Ideker T.2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks.Genome Res, 13(11): 2498-2504. |
| [35] | Song W B, Zhao H M, Zhang X B, Lei L, Lai J S.2016. Genome-wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in maize.Front Plant Sci, 6: 1177. |
| [36] | Turck F, Zhou A, Somssich I E.2004. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related genePcPR1-1 in parsley. Plant Cell, 16(10): 2573-2585. |
| [37] | Ülker B, Somssich I E.2004. WRKY transcription factors: From DNA binding towards biological function.Curr Opin Plant Biol, 7(5): 491-498. |
| [38] | Wang H H, Meng J, Peng X X, Tang X K, Zhou P L, Xiang J H, Deng X B.2015. Rice WRKY4 acts as a transcriptional activator mediating defense responses towardRhizoctonia solani, the causing agent of rice sheath blight. Plant Mol Biol, 89: 157-171. |
| [39] | Wang H P, Hu Y R, Pan J J, Yu D Q.2015. Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci Rep, 5: 14185. |
| [40] | Wang H Z, Avci U, Nakashima J, Hahn M G, Chen F, Dixon R A.2010. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants.Proc Natl Acad Sci USA, 107: 22338-22343. |
| [41] | Wang M, Vannozzi A, Wang G, Zhong Y, Corso M, Cavallini E, Cheng Z M.2015. A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins.Front Plant Sci, 6: 417. |
| [42] | Xiao J, Cheng H T, Li X H, Xiao J H, Xu C G, Wang S P.2013. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements.Plant Physiol, 163(4): 1868-1882. |
| [43] | Xie Y D, Li W, Guo D, Dong J, Zhang Q, Fu Y, Ren D, Peng M, Xia Y.2010. TheArabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate- mediated defence responses. Plant Cell Environ, 33(5): 828-839. |
| [44] | Zhou Y, Yang Y, Zhou X J, Chi Y J, Fan B F, Chen Z X.2016. Structural and functional characterization of the VQ protein family and VQ protein variants from soybean.Sci Rep, 6: 34663. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||