
Rice Science ›› 2025, Vol. 32 ›› Issue (3): 307-321.DOI: 10.1016/j.rsci.2025.01.007
• Reviews • Previous Articles Next Articles
Chen Su1,#, Ma Feilong5,#, Chen Jiaoyang5, Qi Man5, Wei Qianshu1, Tao Zhihuan4( ), Sun Bo2,3(
), Sun Bo2,3( )
)
Received:2024-11-04
Accepted:2025-01-23
Online:2025-05-28
Published:2025-06-16
Contact:
Sun Bo (About author:First author contact: These authors contributed equally to this work
Chen Su, Ma Feilong, Chen Jiaoyang, Qi Man, Wei Qianshu, Tao Zhihuan, Sun Bo. Function of R2R3-Type Myeloblastosis Transcription Factors in Plants[J]. Rice Science, 2025, 32(3): 307-321.
Add to citation manager EndNote|Ris|BibTeX
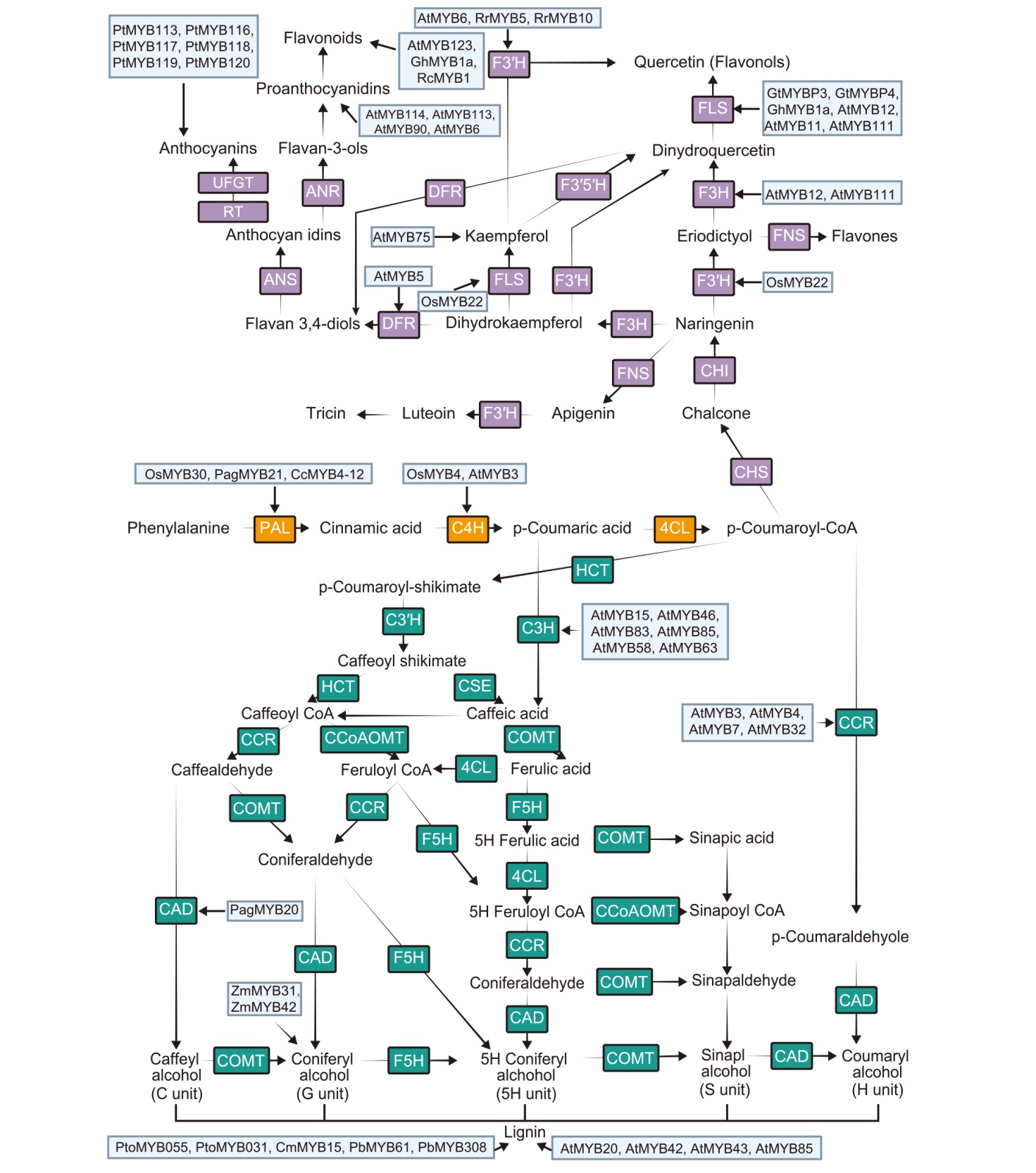
Fig. 1. Tight relationship between myeloblastosis (MYB) and phenylpropanoid metabolism. PAL, Phenylalanine ammonialyase; C4H, Cinnamic acid 4-hydroxylase; 4CL, 4-Coumarate-CoA ligase; CHS, Chalcone synthase; CHI, Chalcone isomerase; F3H, Flavanone 3-hydroxylase; FNS, Flavone synthase; F3′H, Flavonoid 3′-hydroxylase; DFR, Fihydroflavonol 4-reductase; FLS, Flavonol synthase; F3′5′H, Flavonoid 3′5′-hydroxylase; ANR, Anthocyanidin reductase; ANS, Anthocyanin synthase; RT, Rhamnosyl transferase; UFGT, UDPG-flavonoid glucosyltransferase; HCT, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; C3′H, p-coumaroyl shikimate 3′ hydroxylase; C3H, Coumarate 3-hydroxylase; CSE, Caffeoyl shikimate esterase; COMT, Caffeate/5-hydroxyferulate 3-O-methyltransferase; CCoAOMT, Caffeoyl CoA 3-O-methyltransferase; F5H, Ferulate 5-hydroxylase; CAD, Cinnamyl alcohol dehydrogenase; CCR, Cinnamoyl-CoA reductase.
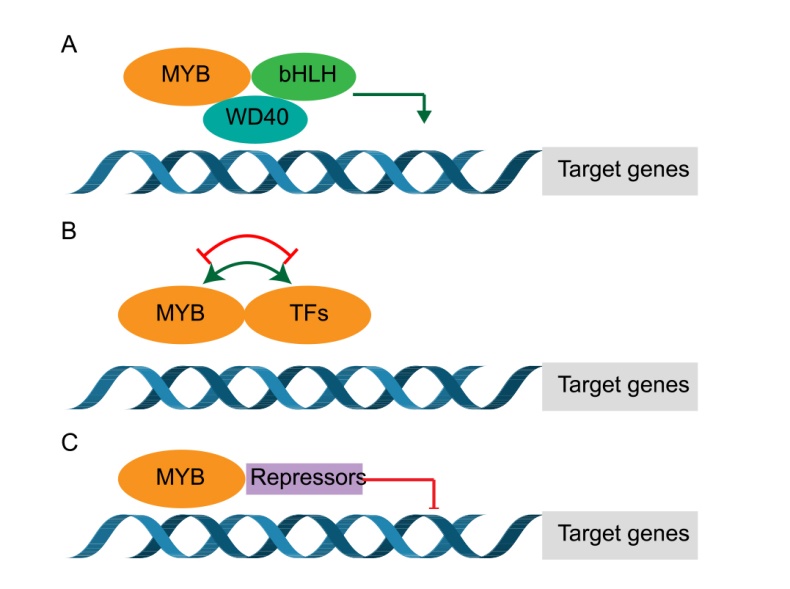
Fig. 2. Schematic model of myeloblastosis (MYB) transcriptional regulation mechanisms. A, MBW (MYB, bHLH, and WD40) complex. B, MYB forms complexes with other transcription factors (TFs). C, Repressors affect the function of MYBs by interacting with them.
| [1] | An C, Sheng L P, Du X P, et al. 2019. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic Res, 6: 84. |
| [2] | An J P, Li R, Qu F J, et al. 2018. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J, 96(3): 562-577. |
| [3] | An J P, Wang X F, Zhang X W, et al. 2020. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J, 18(2): 337-353. |
| [4] | Balhara R, Verma D, Kaur R, et al. 2024. MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea BMC Plant Biol, 24(1): 999. |
| [5] | Bar M, Israeli A, Levy M, et al. 2016. CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell, 28(7): 1602-1615. |
| [6] | Cai H S, Tian S, Liu C L, et al. 2011. Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene, 485(2): 146-152. |
| [7] | Cai Y P, Liu Y T, Fan Y Y, et al. 2023. MYB112 connects light and circadian clock signals to promote hypocotyl elongation in Arabidopsis. Plant Cell, 35(9): 3485-3503. |
| [8] | Chen K Q, Song M R, Guo Y N, et al. 2019. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol J, 17(12): 2341-2355. |
| [9] | Chen L M, Yang H L, Fang Y S, et al. 2021. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol J, 19(4): 702-716. |
| [10] | Chen N, Pan L J, Yang Z, et al. 2023. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways. Front Plant Sci, 14: 1136626. |
| [11] | Chen T Z, Li W J, Hu X H, et al. 2015. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol, 56(5): 917-929. |
| [12] | Chen Y, Wang H Y, Chen Y F. 2021. The transcription factor MYB 40 is a central regulator in arsenic resistance in Arabidopsis. Plant Commun, 2(6): 100234. |
| [13] | Coleto I, Bejarano I, Marín-Peña A J, et al. 2021. Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol, 229(2): 1021-1035. |
| [14] | Cui F Q, Li X X, Wu W W, et al. 2022.Ectopic expression of BOTRYTIS SUSCEPTIBLE1 reveals its function as a positive regulator of wound-induced cell death and plant susceptibility to Botrytis . Plant Cell, 34(10): 4105-4116. |
| [15] | Cui J, Jiang N, Zhou X X, et al. 2018. Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta, 248(6): 1487-1503. |
| [16] | Dai X Y, Wang Y Y, Yang A, et al. 2012. OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol, 159(1): 169-183. |
| [17] | de Vos M, Denekamp M, Dicke M, et al. 2006. The Arabidopsis thaliana transcription factor AtMYB 102 functions in defense against the insect herbivore Pieris rapae. Plant Signal Behav, 1(6): 305-311. |
| [18] | Ding A M, Tang X F, Yang D H, et al. 2021. ERF4 and MYB52 transcription factors play antagonistic roles in regulating homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage. Plant Cell, 33(2): 381-403. |
| [19] | Ding B Q, Patterson E L, Holalu S V, et al. 2020. Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr Biol, 30(5): 802-814. |
| [20] | Dubos C, Stracke R, Grotewold E, et al. 2010. MYB transcription factors in Arabidopsis. Trends Plant Sci, 15(10): 573-581. |
| [21] | Elomaa P, Uimari A, Mehto M, et al. 2003. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol, 133(4): 1831-1842. |
| [22] | Feng S Q, Wang Y L, Yang S, et al. 2010. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10 Planta, 232(1): 245-255. |
| [23] | Fornalé S, Shi X H, Chai C L, et al. 2010. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J, 64(4): 633-644. |
| [24] | Geng D L, Chen P X, Shen X X, et al. 2018. MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol, 178(3): 1296-1309. |
| [25] | Geng P, Zhang S, Liu J Y, et al. 2020. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol, 182(3): 1272-1283. |
| [26] | Gonzalez A, Zhao M Z, Leavitt J M, et al. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J, 53(5): 814-827. |
| [27] | Gu K D, Zhang Q Y, Yu J Q, et al. 2021. R2R3-MYB transcription factor MdMYB73 confers increased resistance to the fungal pathogen Botryosphaeria dothidea in apples via the salicylic acid pathway. J Agric Food Chem, 69(1): 447-458. |
| [28] | Gui J S, Luo L F, Zhong Y, et al. 2019. Phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol Plant, 12(10): 1325-1337. |
| [29] | He G R, Zhang R, Jiang S H, et al. 2023. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hortic Res, 10(6): uhad080. |
| [30] | He J, Liu Y Q, Yuan D Y, et al. 2020. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci USA, 117(1): 271-277. |
| [31] | Heim M A, Jakoby M, Werber M, et al. 2003. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol, 20(5): 735-747. |
| [32] | Huang C J, Hu G J, Li F F, et al. 2013. NbPHAN, a MYB transcriptional factor, regulates leaf development and affects drought tolerance in Nicotiana benthamiana Physiol Plant, 149(3): 297-309. |
| [33] | Huang H, Gong Y L, Liu B, et al. 2020. The DELLA proteins interact with MYB21 and MYB 24 to regulate filament elongation in Arabidopsis. BMC Plant Biol, 20(1): 64. |
| [34] | Huang W J, Sun W, Lv H Y, et al. 2013. A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLoS One, 8(8): e70778. |
| [35] | Huang Y J, Zhao H X, Gao F, et al. 2018. A R2R3-MYB transcription factor gene, FtMYB13, from Tartary buckwheat improves salt/ drought tolerance in Arabidopsis. Plant Physiol Biochem, 132: 238-248. |
| [36] | Im J H, Son S, Kim W C, et al. 2024. Jasmonate activates secondary cell wall biosynthesis through MYC2-MYB46 module. Plant J, 117(4): 1099-1114. |
| [37] | Jahan M A, Harris B, Lowery M, et al. 2020. Glyceollin transcription factor GmMYB29A 2 regulates soybean resistance to Phytophthora sojae. Plant Physiol, 183(2): 530-546. |
| [38] | Kang Y H, Song S K, Schiefelbein J, et al. 2013. Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol, 163(1): 193-204. |
| [39] | Kaur H, Heinzel N, Schöttner M, et al. 2010. R2R3-NaMYB 8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol, 152(3): 1731-1747. |
| [40] | Kim D, Jeon S J, Yanders S, et al. 2022. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep, 41(7): 1549-1560. |
| [41] | Kim S H, Lam P Y, Lee M H, et al. 2020. The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front Plant Sci, 11: 583153. |
| [42] | Legay S, Lacombe E, Goicoechea M, et al. 2007. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci, 173(5): 542-549. |
| [43] | Li W T, Zhu Z W, Chern M, et al. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell, 170(1): 114-126. |
| [44] | Li W T, Wang K, Chern M, et al. 2020. Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol, 226(6): 1850-1863. |
| [45] | Li Y, Hu Z Y, Dong Y M, et al. 2023. Overexpression of the cotton trihelix transcription factor GhGT23 in Arabidopsis mediates salt and drought stress tolerance by binding to GT and MYB promoter elements in stress-related genes. Front Plant Sci, 14: 1144650. |
| [46] | Li Y Q, Shan X T, Tong L N, et al. 2020. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis. Plant Cell Physiol, 61(7): 1365-1380. |
| [47] | Liao K, Peng Y J, Yuan L B, et al. 2020. Brassinosteroids antagonize jasmonate-activated plant defense responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol, 182(2): 1066-1082. |
| [48] | Liao Y, Zou H F, Wang H W, et al. 2008. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res, 18(10): 1047-1060. |
| [49] | Lin H, Wang M Y, Chen Y, et al. 2022. An MKP-MAPK protein phosphorylation cascade controls vascular immunity in plants. Sci Adv, 8(10): eabg8723. |
| [50] | Liu H N, Shu Q, Kui L W, et al. 2021. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol Hortic, 1(1): 14. |
| [51] | Liu H X, Zhou X Y, Dong N, et al. 2011. Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Funct Integr Genomics, 11(3): 431-443. |
| [52] | Liu T L, Chen T Z, Kan J L, et al. 2022. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol J, 20(4): 722-735. |
| [53] | Liu Y H, Kui L W, Espley R V, et al. 2019. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in Tuber flesh of potato. J Exp Bot, 70(15): 3809-3824. |
| [54] | Lloyd A, Brockman A, Aguirre L, et al. 2017. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol, 58(9): 1431-1441. |
| [55] | Luo G B, Shen L S, Song Y H, et al. 2021. The MYB family transcription factor TuODORANT1 from Triticum urartu and the homolog TaODORANT1 from Triticum aestivum inhibit seed storage protein synthesis in wheat. Plant Biotechnol J, 19(9): 1863-1877. |
| [56] | Meng Y Y, Wang Z Y, Wang Y Q, et al. 2019.The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell, 31(11): 2751-2767. |
| [57] | Michaletz S, Cheng D, Kerkhoff A, et al. 2014. Convergence of terrestrial plant production across global climate gradients. Nature, 512: 39-43. |
| [58] | Morse A M, Whetten R W, Dubos C, et al. 2009. Post-translational modification of an R2R3-MYB transcription factor by a MAP Kinase during xylem development. New Phytol, 183(4): 1001-1013. |
| [59] | Nakatsuka T, Saito M, Yamada E, et al. 2012. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot, 63(18): 6505-6517. |
| [60] | Nguyen N H, Cheong J J. 2018. AtMYB44 interacts with TOPLESS-RELATED corepressors to suppress protein phosphatase 2C gene transcription. Biochem Biophys Res Commun, 507(1/4): 437-442. |
| [61] | Ni J B, Premathilake A T, Gao Y H, et al. 2021. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J, 105(1): 167-181. |
| [62] | Noman A, Hussain A, Adnan M, et al. 2019. A novel MYB transcription factor CaPHL8 provide clues about evolution of pepper immunity against soil borne pathogen. Microb Pathog, 137: 103758. |
| [63] | Onkokesung N, Reichelt M, van Doorn A, et al. 2014. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J Exp Bot, 65(8): 2203-2217. |
| [64] | Pandey A, Misra P, Trivedi P K. 2015. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep, 34(9):1515-1528. |
| [65] | Palapol Y, Ketsa S, Kui L W, et al. 2009. A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta, 229(6): 1323-1334. |
| [66] | Park J S, Kim J B, Cho K J, et al. 2008. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep, 27(6): 985-994. |
| [67] | Pattanaik S, Kong Q, Zaitlin D, et al. 2010. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta, 231(5): 1061-1076. |
| [68] | Paz-Ares J, Ghosal D, Wienand U, et al. 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J, 6(12): 3553-3558. |
| [69] | Qin Y X, Wang M C, Tian Y C, et al. 2012. Over-expression of TaMYB 33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol Biol Rep, 39(6): 7183-7192. |
| [70] | Qiu J H, Xie J H, Chen Y, et al. 2022. Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice. Mol Plant, 15(4): 723-739. |
| [71] | Qiu Y P, Tao R, Feng Y, et al. 2021. EIN3 and RSL4 interfere with an MYB-bHLH-WD40 complex to mediate ethylene-induced ectopic root hair formation in Arabidopsis Proc Natl Acad Sci USA, 118(51): e2110004118. |
| [72] | Qiu Z K, Yan S S, Xia B, et al. 2019. The eggplant transcription factor MYB44 enhances resistance to bacterial wilt by activating the expression of spermidine synthase. J Exp Bot, 70(19): 5343-5354. |
| [73] | Ramsay N A, Glover B J. 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci, 10(2): 63-70. |
| [74] | Ren D Y, Rao Y C, Yu H P, et al. 2020. MORE FLORET1 encodes a MYB transcription factor that regulates spikelet development in rice. Plant Physiol, 184(1): 251-265. |
| [75] | Schiefelbein J, Kwak S H, Wieckowski Y, et al. 2009. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis J Exp Bot, 60(5): 1515-1521. |
| [76] | Serpa V, Vernal J, Lamattina L, et al. 2007. Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem Biophys Res Commun, 361(4): 1048-1053. |
| [77] | Shan H, Chen S M, Jiang J F, et al. 2012.Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana Mol Biotechnol, 51(2): 160-173. |
| [78] | Sharma N, Sahu P P, Prasad A, et al. 2021. The Sw5a gene confers resistance to ToLCNDV and triggers an HR response after direct AC4 effector recognition. Proc Natl Acad Sci USA, 118(33): e2101833118. |
| [79] | Sharma S B, Dixon R A. 2005.Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana Plant J, 44(1): 62-75. |
| [80] | Shen Y X, Sun T T, Pan Q, et al. 2019. RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa Plant Biotechnol J, 17(11): 2078-2095. |
| [81] | Song S, Guo W, Guo Y, et al. 2024. Transcription factor PdMYB118 in poplar regulates lignin deposition and xylem differentiation in addition to anthocyanin synthesis through suppressing the expression of PagKNAT2/6b gene. Plant Sci, 350: 112277. |
| [82] | Song S S, Qi T C, Huang H, et al. 2011. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB 24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell, 23(3): 1000-1013. |
| [83] | Sun B, Shen Y J, Chen S, et al. 2023. A novel transcriptional repressor complex MYB22-TOPLESS-HDAC1 promotes rice resistance to brown planthopper by repressing F3’H expression. New Phytol, 239(2): 720-738. |
| [84] | Sun C, Wang C M, Zhang W, et al. 2021. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic Res, 8(1): 156. |
| [85] | Sun Y M, Ren S, Ye S L, et al. 2020. Identification and functional characterization of PtoMYB 055 involved in the regulation of the lignin biosynthesis pathway in Populus tomentosa. Int J Mol Sci, 21(14): 4857. |
| [86] | Tamagnone L, Merida A, Parr A, et al. 1998. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell, 10(2): 135-154. |
| [87] | Tan H J, Man C, Xie Y, et al. 2019. A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis Mol Plant, 12(4): 521-537. |
| [88] | Tang Y H, Bao X X, Zhi Y L, et al. 2019. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front Plant Sci, 10: 168. |
| [89] | Wan J P, Wang R L, Zhang P, et al. 2021. MYB70 modulates seed germination and root system development in Arabidopsis. iScience, 24(11): 103228. |
| [90] | Wang B H, Luo Q, Li Y P, et al. 2022. Structural insights into partner selection for MYB and bHLH transcription factor complexes. Nat Plants, 8(9): 1108-1117. |
| [91] | Wang H, Zhang H, Yang Y, et al. 2020. The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × Ananassa) fruits. Plant Biotechnol J, 18(5): 1169-1184. |
| [92] | Wang H, Wang Z X, Tian H Y, et al. 2024. The miR172a-SNB module orchestrates both induced and adult-plant resistance to multiple diseases via MYB30-mediated lignin accumulation in rice. Mol Plant, S1674- 2052(24)00383-6. |
| [93] | Wang H Z, Yang K Z, Zou J J, et al. 2015. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat Commun, 6: 8822. |
| [94] | Wang J, Nan N, Li N, et al. 2020. A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell, 32(11): 3535-3558. |
| [95] | Wang L J, Lu W X, Ran L Y, et al. 2019. R2R3-MYB transcription factor MYB 6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J, 99(4): 733-751. |
| [96] | Wang X, Ding Y L, Li Z Y, et al. 2019. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15 Dev Cell, 51(2): 222-235. |
| [97] | Wang Z D, Li X X, Yao X H, et al. 2023. MYB44 regulates PTI by promoting the expression of EIN2 and MPK3/6 in Arabidopsis. Plant Commun, 4(6): 100628. |
| [98] | Wu M B, Zhou Y Y, Ma H F, et al. 2024. SlMYB72 interacts with SlTAGL1 to regulate the cuticle formation in tomato fruit. Plant J, 120(4): 1591-1604. |
| [99] | Xiao S H, Hu Q, Shen J L, et al. 2021. GhMYB 4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae. Plant Cell Rep, 40(4): 735-751. |
| [100] | Xie L H, Yan T X, Li L, et al. 2021. An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua New Phytol, 231(5): 2050-2064. |
| [101] | Xie Y P, Chen P X, Yan Y, et al. 2018. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol, 218(1): 201-218. |
| [102] | Xiong C, Xie Q M, Yang Q H, et al. 2020. WOOLLY, interacting with MYB transcription factor MYB31, regulates cuticular wax biosynthesis by modulating CER6 expression in tomato. Plant J, 103(1): 323-337. |
| [103] | Xu W J, Grain D, Bobet S, et al. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol, 202(1): 132-144. |
| [104] | Xu W J, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci, 20(3): 176-185. |
| [105] | Yan H L, Pei X N, Zhang H, et al. 2021. MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci, 22(6): 3103. |
| [106] | Yan Y, Li C, Dong X J, et al. 2020. MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell, 32(7): 2196-2215. |
| [107] | Yang C H, Li D Y, Liu X, et al. 2014. OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol, 14: 158. |
| [108] | Yang X, Li X, Shan J M, et al. 2021. Overexpression of GmGAMYB accelerates the transition to flowering and increases plant height in soybean. Front Plant Sci, 12: 667242. |
| [109] | Yang X H, Luo Y C, Bai H R, et al. 2022. DgMYB2 improves cold resistance in chrysanthemum by directly targeting DgGPX1. Hortic Res, 9: uhab028. |
| [110] | Yang Y, Zhang L B, Chen P, et al. 2020. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J, 39(2): e101928. |
| [111] | Yang Z Z, Li Y Q, Gao F Z, et al. 2020. MYB 21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J Exp Bot, 71(14): 4140-4158. |
| [112] | Yao G F, Ming M L, Allan A C, et al. 2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J, 92(3): 437-451. |
| [113] | Yu S, Zhang W, Zhang L P, et al. 2024. Negative regulation of CcPAL2 gene expression by the repressor transcription factor CcMYB4-12 modulates lignin and capsaicin biosynthesis in Capsicum chinense fruits. Int J Biol Macromol, 280: 135592. |
| [114] | Zhang C Y, Liu H C, Zhang X S, et al. 2020. VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosum, negatively regulates salt, drought, and temperature stress. Gene, 757: 144935. |
| [115] | Zhang L C, Zhao G Y, Xia C, et al. 2012. Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene, 505(1): 100-107. |
| [116] | Zhang M F, Wang J Q, Luo Q J, et al. 2021. CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation. J Plant Physiol, 264: 153472. |
| [117] | Zhang P, Wang R L, Ju Q, et al. 2019. The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol, 180(1): 529-542. |
| [118] | Zhang S C, Ma P D, Yang D F, et al. 2013. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS One, 8(9): e73259. |
| [119] | Zhang Y H, Chen S, Xu L H, et al. 2024. Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar. Plant Cell, 36(5): 1806-1828. |
| [120] | Zhang Z Y, Liu X, Wang X D, et al. 2012. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol, 196(4): 1155-1170. |
| [121] | Zhao H X, Yao P F, Zhao J L, et al. 2022. A novel R2R3-MYB transcription factor FtMYB22 negatively regulates salt and drought stress through ABA-dependent pathway. Int J Mol Sci, 23(23): 14549. |
| [122] | Zheng H Y, Dong L L, Han X Y, et al. 2020. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol, 225(6): 2526-2541. |
| [123] | Zhong C M, Tang Y, Pang B, et al. 2020. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic Res, 7: 78. |
| [124] | Zhou M L, Sun Z M, Ding M Q, et al. 2017a. FtSAD2 and FtJAZ 1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol, 216(3): 814-828. |
| [125] | Zhou M L, Zhang K X, Sun Z M, et al. 2017b. LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol, 174(3): 1348-1358. |
| [126] | Zhu S J, Shi W J, Jie Y C, et al. 2020. A MYB transcription factor, BnMYB2, cloned from ramie (Boehmeria nivea) is involved in cadmium tolerance and accumulation. PLoS One, 15(5): e0233375. |
| [127] | Zhu Y S, Wang Y C, Jiang H Y, et al. 2023. Transcriptome analysis reveals that PbMYB61 and PbMYB308 are involved in the regulation of lignin biosynthesis in pear fruit stone cells. Plant J, 116(1): 217-233. |
| [128] | Zhu Y T, Hu X Q, Wang P, et al. 2022. GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway. Int J Biol Macromol, 201: 580-591. |
| [1] | Yang Yajun, Lu Yanhui, Tian Junce, Zheng Xusong, Guo Jiawen, Liu Xiaowei, Lü Zhongxian, Xu Hongxing. Sustainable Management Strategies for Rice Leaffolder, Cnaphalocrocis medinalis (Guenée): Progress and Prospects [J]. Rice Science, 2025, 32(3): 322-338. |
| [2] | Xie Yuhao, Xie Wenya, Zhao Jianhua, Xue Xiang, Cao Wenlei, Shi Xiaopin, Wang Zhou, Wang Yiwen, Wang Guangda, Feng Zhiming, Hu Keming, Chen Xijun, Chen Zongxiang, Zuo Shimin. OsERF7 Negatively Regulates Resistance to Sheath Blight Disease by Inhibiting Phytoalexin Biosynthesis [J]. Rice Science, 2025, 32(3): 367-379. |
| [3] | Chaemyeong Lim, Sae Hyun Lee, Haeun Lee, So-Yon Park, Kiyoon Kang, Hyeryung Yoon, Tae-Jin Yang, Gary Stacey, Nam-Chon Paek, Sung-Hwan Cho. Global Transcriptome Analysis of Rice Seedlings in Response to Extracellular ATP [J]. Rice Science, 2025, 32(3): 380-399. |
| [4] | Zeng Deyong, Cui Jie, Yin Yishu, Dai Cuihong, Yu Wencheng, Zhao Haitian, Guan Shuanghong, Cheng Dayou, Sun Yeqing, Lu Weihong. Generational Genetic Mechanism of Space Mutagenesis in Rice Based on Multi-Omics [J]. Rice Science, 2025, 32(3): 400-425. |
| [5] | Sanchika Snehi, Ravi Kiran Kt, Sanket Rathi, Sameer Upadhyay, Suneetha Kota, Satish Kumar Sanwal, Lokeshkumar Bm, Arun Balasubramaniam, Nitish Ranjan Prakash, Pawan Kumar Singh. Discerning Genes to Deliver Varieties: Enhancing Vegetative- and Reproductive-Stage Flooding Tolerance in Rice [J]. Rice Science, 2025, 32(2): 160-176. |
| [6] | Nie Lixiao, Guo Xiayu, Wang Weiqin, Qi Yucheng, Ai Zhiyong, He Aibin. Regulation of Regeneration Rate to Enhance Ratoon Rice Production [J]. Rice Science, 2025, 32(2): 177-192. |
| [7] | Wang Shuman, Zhang Linqi, Gao Ruiren, Wei Guangbo, Dong Weiguo, Xu Jiming, Wang Zhiye. Establishing Programmable CRISPR/Cas13b-Mediated Knockdown System in Rice [J]. Rice Science, 2025, 32(2): 217-227. |
| [8] | Uthpal Krishna Roy, Babita Pal, Soumen Bhattacharjee. A Novel Approach for Screening Salinity-Tolerant Rice Germplasm by Exploring Redox-Regulated Cytological Fingerprint [J]. Rice Science, 2025, 32(2): 228-242. |
| [9] | He Zhenrui, Zhao Wenhua, Cheng Baoping, Yang Mei, Yang Yingqing, Zhu Yiming, Zhou Erxun. Molecular and Biological Characterization of Novel Mitovirus Infecting Phytopathogenic Fungus Ustilaginoidea virens [J]. Rice Science, 2025, 32(2): 243-258. |
| [10] | He Chen, Ruan Yunze, Jia Zhongjun. A Meta-Analysis of 30 Years in China and Micro-District Experiments Shows Organic Fertilizer Quantification Combined with Chemical Fertilizer Reduction Enhances Rice Yield on Saline-Alkali Land [J]. Rice Science, 2025, 32(2): 259-272. |
| [11] | Wang Mingyue, Zhao Weibo, Feng Xiaoya, Chen Yi, Li Junhao, Fu Jinmei, Yan Yingchun, Chu Zhaohui, Huang Wenchao. Disruption of Energy Metabolism and Reactive Oxygen Species Homeostasis in Honglian Type-Cytoplasmic Male Sterility (HL-CMS) Rice Pollen [J]. Rice Science, 2025, 32(1): 81-93. |
| [12] | Intan Farahanah, Shariza Sahudin, Hannis Fadzillah Mohsin, Siti Alwani Ariffin, Liyana Dhamirah Aminuddin. Understanding Investigational Perspective of Antioxidant and Antibacterial Properties of Rice [J]. Rice Science, 2025, 32(1): 15-31. |
| [13] | Jeberlin Prabina Bright, Hemant S. Maheshwari, Sugitha Thangappan, Kahkashan Perveen, Najat A. Bukhari, Debasis Mitra, Riyaz Sayyed, Andrea Mastinu. Biofilmed-PGPR: Next-Generation Bioinoculant for Plant Growth Promotion in Rice under Changing Climate [J]. Rice Science, 2025, 32(1): 94-106. |
| [14] | Wang Haoran, Chen Guoqing, Feng Guozhong. Expanding Viral Diversity in Rice Fields by Next-Generation Sequencing [J]. Rice Science, 2025, 32(1): 44-51. |
| [15] | Surangkana Chimthai, Sulaiman Cheabu, Wanchana Aesomnuk, Siriphat Ruengphayak, Siwaret Arikit, Apichart Vanavichit, Chanate Malumpong. Breeding for Heat Tolerant Aromatic Rice Varieties and Identification of Novel QTL Regions Associated with Heat Tolerance During Reproductive Phase by QTL-Seq [J]. Rice Science, 2025, 32(1): 67-80. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||