
Rice Science ›› 2019, Vol. 26 ›› Issue (1): 11-20.DOI: 10.1016/j.rsci.2018.12.001
• Orginal Article • Previous Articles Next Articles
Lei He, Guang Chen, Sen Zhang, Zhennan Qiu, Jiang Hu, Dali Zeng, Guangheng Zhang, Guojun Dong, Zhenyu Gao, Deyong Ren, Lan Shen, Longbiao Guo, Qian Qian( ), Li Zhu(
), Li Zhu( )
)
Received:2018-05-26
Accepted:2018-07-31
Online:2019-01-29
Published:2018-10-22
Lei He, Guang Chen, Sen Zhang, Zhennan Qiu, Jiang Hu, Dali Zeng, Guangheng Zhang, Guojun Dong, Zhenyu Gao, Deyong Ren, Lan Shen, Longbiao Guo, Qian Qian, Li Zhu. Functional Analysis of Three Rice Chloroplast Transit Peptides[J]. Rice Science, 2019, 26(1): 11-20.
Add to citation manager EndNote|Ris|BibTeX
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::CTPTRXz-GFP-F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCC | CTPTRXz GFP | 168 |
| p35S::CTPTRXz-GFP-R | ATGGATCCTCTAGAGTCGACGCGACACCCACCGAGGAT | ||
| p35S::CTPHSA1-GFP-F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | CTPHSA1 GFP | 162 |
| p35S::CTPHSA1-GFP-R | ATGGATCCTCTAGAGTCGACACTTACGTTTCGGTTGAG | ||
| p35S::36CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCC | CTPFLN1 GFP | 108 |
| p35S::36CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACGCGAATATGGCGGCCTCG |
Table 1 Nucleotide sequences used in construction of fusion vector with chloroplast transit peptides and GFP (green fluorescent protein).
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::CTPTRXz-GFP-F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCC | CTPTRXz GFP | 168 |
| p35S::CTPTRXz-GFP-R | ATGGATCCTCTAGAGTCGACGCGACACCCACCGAGGAT | ||
| p35S::CTPHSA1-GFP-F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | CTPHSA1 GFP | 162 |
| p35S::CTPHSA1-GFP-R | ATGGATCCTCTAGAGTCGACACTTACGTTTCGGTTGAG | ||
| p35S::36CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCC | CTPFLN1 GFP | 108 |
| p35S::36CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACGCGAATATGGCGGCCTCG |
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::ΔCTPTRXz-GFP-F | TATTTACAATTACAGTCGACATGGCGCAAGGCGTTCGA | ΔCTPTRXz GFP | 402 |
| p35S::ΔCTPTRXz-GFP-R | ATGGATCCTCTAGAGTCGACCAATTCATTATCAATGAT | ||
| p35S::Δ36CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGTGCTCCCCGAACGGC | Δ36CTPFLN1 GFP | 1 488 |
| p35S::Δ36CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA | ||
| p35S::Δ50CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGGCCCCACGCCGCGGG | Δ50CTPFLN1 GFP | 1 446 |
| p35S::Δ50CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA | ||
| p35S::Δ89CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGCCGAAGCGGCGGGGC | Δ89CTPFLN1 GFP | 1 329 |
| p35S::Δ89CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA |
Table 2 Nucleotide sequences used in construction of GFP (green fluorescent protein) fusion vector for deletion of chloroplast transit peptide.
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::ΔCTPTRXz-GFP-F | TATTTACAATTACAGTCGACATGGCGCAAGGCGTTCGA | ΔCTPTRXz GFP | 402 |
| p35S::ΔCTPTRXz-GFP-R | ATGGATCCTCTAGAGTCGACCAATTCATTATCAATGAT | ||
| p35S::Δ36CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGTGCTCCCCGAACGGC | Δ36CTPFLN1 GFP | 1 488 |
| p35S::Δ36CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA | ||
| p35S::Δ50CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGGCCCCACGCCGCGGG | Δ50CTPFLN1 GFP | 1 446 |
| p35S::Δ50CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA | ||
| p35S::Δ89CTPFLN1-GFP-F | TATTTACAATTACAGTCGACATGCCGAAGCGGCGGGGC | Δ89CTPFLN1 GFP | 1 329 |
| p35S::Δ89CTPFLN1-GFP-R | ATGGATCCTCTAGAGTCGACCCACATAGAAGGCACATA |
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::CTPTRXz-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-Ghd10-GFP-1R | GGCTCCTGCGCCGCCGCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-Ghd10-GFP-2F | CATCCTCGGTGGGTGTCGCATGGCGGCGGCGCAGGAGCC | Ghd10 full length | 1 230 |
| p35S::CTPTRXz-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::CTPTRXz-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-SUI1-GFP-1R | GATGACCATTGACCTCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-SUI1-GFP-2F | CATCCTCGGTGGGTGTCGCATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::CTPTRXz-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::CTPTRXz-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-MFS1-GFP-1R | CTCCCATGTCCAGCTCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-MFS1-GFP-2F | CATCCTCGGTGGGTGTCGCATGGAGCTGGACATGGGAG | MFS1 full length | 570 |
| p35S::CTPTRXz-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::36CTPFLN1-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-Ghd10-GFP-1R | GGCTCCTGCGCCGCCGCCATGCGAATATGGCGGCCTCGG | ||
| p35S::36CTPFLN1-Ghd10-GFP-2F | CCGAGGCCGCCATATTCGCATGGCGGCGGCGCAGGAGCC | Ghd10 full length | 1 230 |
| p35S::36CTPFLN1-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::36CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-SUI1-GFP-1R | CCGAGGCCGCCATATTCGCATGGAGGTCAATGGTCATC | ||
| p35S::36CTPFLN1-SUI1-GFP-2F | GATGACCATTGACCTCCATGCGAATATGGCGGCCTCGG | SUI1 full length | 1 278 |
| p35S::36CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::36CTPFLN1-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-MFS1-GFP-1R | CCGAGGCCGCCATATTCGCATGGAGCTGGACATGGGAG | ||
| p35S::36CTPFLN1-MFS1-GFP-2F | CTCCCATGTCCAGCTCCATGCGAATATGGCGGCCTCGG | MFS1 full length | 570 |
| p35S::36CTPFLN1-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::CTPHSA1-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-Ghd10-GFP-1R | GCTCAACCGAAACGTAAGTATGGCGGCGGCGCAGGAGC | ||
| p35S::CTPHSA1-Ghd10-GFP-2F | GCTCCTGCGCCGCCGCCATACTTACGTTTCGGTTGAGC | Ghd10 full length | 1 230 |
| p35S::CTPHSA1-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::CTPHSA1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-SUI1-GFP-1R | GCTCAACCGAAACGTAAGTATGGAGGTCAATGGTCATC | ||
| p35S::CTPHSA1-SUI1-GFP-2F | GATGACCATTGACCTCCATACTTACGTTTCGGTTGAGC | SUI1 full length | 1 278 |
| p35S::CTPHSA1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::CTPHSA1-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-MFS1-GFP-1R | GCTCAACCGAAACGTAAGTATGGAGCTGGACATGGGAG | ||
| p35S::CTPHSA1-MFS1-GFP-2F | CTCCCATGTCCAGCTCCATACTTACGTTTCGGTTGAGC | MFS1 full length | 570 |
| p35S::CTPHSA1-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::50CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 50CTP | 150 |
| p35S::50CTPFLN1-SUI1-GFP-1R | GATGACCATTGACCTCCATGGGTTCAGGGGATTCCGGT | ||
| p35S::50CTPFLN1-SUI1-GFP-2F | ACCGGAATCCCCTGAACCCATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::50CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::89CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 89CTP | 267 |
| p35S::89CTPFLN1-SUI1-GFP-1R | GATGACCATTGACCTCCATCGGCTCCTCCTCGCCCTCT | ||
| p35S::89CTPFLN1-SUI1-GFP-2F | AGAGGGCGAGGAGGAGCCGATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::89CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT |
Table 3 Nucleotide sequences used in construction of GFP fusion vector with non-chloroplast-localized gene and chloroplast transit peptide.
| Primer | Sequence (5′-3′) | Experiment | Product length (bp) |
|---|---|---|---|
| p35S::CTPTRXz-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-Ghd10-GFP-1R | GGCTCCTGCGCCGCCGCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-Ghd10-GFP-2F | CATCCTCGGTGGGTGTCGCATGGCGGCGGCGCAGGAGCC | Ghd10 full length | 1 230 |
| p35S::CTPTRXz-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::CTPTRXz-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-SUI1-GFP-1R | GATGACCATTGACCTCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-SUI1-GFP-2F | CATCCTCGGTGGGTGTCGCATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::CTPTRXz-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::CTPTRXz-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCCGCGGCCG | TRXz CTP | 168 |
| p35S::CTPTRXz-MFS1-GFP-1R | CTCCCATGTCCAGCTCCATGCGACACCCACCGAGGATG | ||
| p35S::CTPTRXz-MFS1-GFP-2F | CATCCTCGGTGGGTGTCGCATGGAGCTGGACATGGGAG | MFS1 full length | 570 |
| p35S::CTPTRXz-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::36CTPFLN1-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-Ghd10-GFP-1R | GGCTCCTGCGCCGCCGCCATGCGAATATGGCGGCCTCGG | ||
| p35S::36CTPFLN1-Ghd10-GFP-2F | CCGAGGCCGCCATATTCGCATGGCGGCGGCGCAGGAGCC | Ghd10 full length | 1 230 |
| p35S::36CTPFLN1-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::36CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-SUI1-GFP-1R | CCGAGGCCGCCATATTCGCATGGAGGTCAATGGTCATC | ||
| p35S::36CTPFLN1-SUI1-GFP-2F | GATGACCATTGACCTCCATGCGAATATGGCGGCCTCGG | SUI1 full length | 1 278 |
| p35S::36CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::36CTPFLN1-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 CTP | 108 |
| p35S::36CTPFLN1-MFS1-GFP-1R | CCGAGGCCGCCATATTCGCATGGAGCTGGACATGGGAG | ||
| p35S::36CTPFLN1-MFS1-GFP-2F | CTCCCATGTCCAGCTCCATGCGAATATGGCGGCCTCGG | MFS1 full length | 570 |
| p35S::36CTPFLN1-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::CTPHSA1-Ghd10-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-Ghd10-GFP-1R | GCTCAACCGAAACGTAAGTATGGCGGCGGCGCAGGAGC | ||
| p35S::CTPHSA1-Ghd10-GFP-2F | GCTCCTGCGCCGCCGCCATACTTACGTTTCGGTTGAGC | Ghd10 full length | 1 230 |
| p35S::CTPHSA1-Ghd10-GFP-2R | ATGGATCCTCTAGAGTCGACGAAGTTGTGGCTCCACGTC | ||
| p35S::CTPHSA1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-SUI1-GFP-1R | GCTCAACCGAAACGTAAGTATGGAGGTCAATGGTCATC | ||
| p35S::CTPHSA1-SUI1-GFP-2F | GATGACCATTGACCTCCATACTTACGTTTCGGTTGAGC | SUI1 full length | 1 278 |
| p35S::CTPHSA1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::CTPHSA1-MFS1-GFP-1F | TATTTACAATTACAGTCGACATGCACCGAATGGCTTCT | HSA1 CTP | 162 |
| p35S::CTPHSA1-MFS1-GFP-1R | GCTCAACCGAAACGTAAGTATGGAGCTGGACATGGGAG | ||
| p35S::CTPHSA1-MFS1-GFP-2F | CTCCCATGTCCAGCTCCATACTTACGTTTCGGTTGAGC | MFS1 full length | 570 |
| p35S::CTPHSA1-MFS1-GFP-2R | ATGGATCCTCTAGAGTCGACCGAGCCGAACAGCAGCGCG | ||
| p35S::50CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 50CTP | 150 |
| p35S::50CTPFLN1-SUI1-GFP-1R | GATGACCATTGACCTCCATGGGTTCAGGGGATTCCGGT | ||
| p35S::50CTPFLN1-SUI1-GFP-2F | ACCGGAATCCCCTGAACCCATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::50CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT | ||
| p35S::89CTPFLN1-SUI1-GFP-1F | TATTTACAATTACAGTCGACATGGCCATGGCGGCCTCCC | FLN1 89CTP | 267 |
| p35S::89CTPFLN1-SUI1-GFP-1R | GATGACCATTGACCTCCATCGGCTCCTCCTCGCCCTCT | ||
| p35S::89CTPFLN1-SUI1-GFP-2F | AGAGGGCGAGGAGGAGCCGATGGAGGTCAATGGTCATC | SUI1 full length | 1 278 |
| p35S::89CTPFLN1-SUI1-GFP-2R | ATGGATCCTCTAGAGTCGACTAGCCTTTTCCTTATCATT |
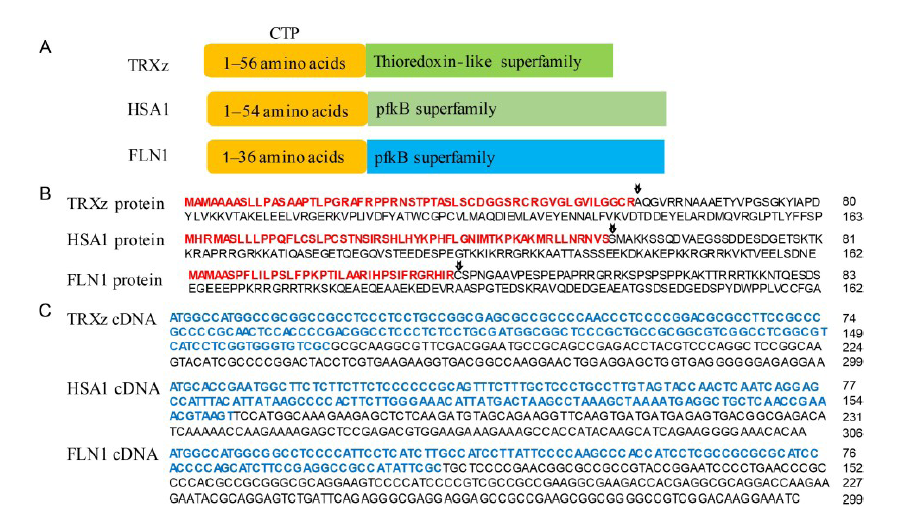
Fig. 1. Structure and sequence of FLN1, HSA1 and TRXz. A, Abbreviated diagram of TRXz, HSA1 and FLN1 protein sequences. B, Amino acids sequences of the three proteins. Sequences of the chloroplast transit peptide (CTP) are shown in red bold type. The arrows indicate the putative cleavage site during chloroplast import based on the TargetP website. C, cDNA sequences encoding the three genes. Sequences of the three CTPs are shown in blue bold type.
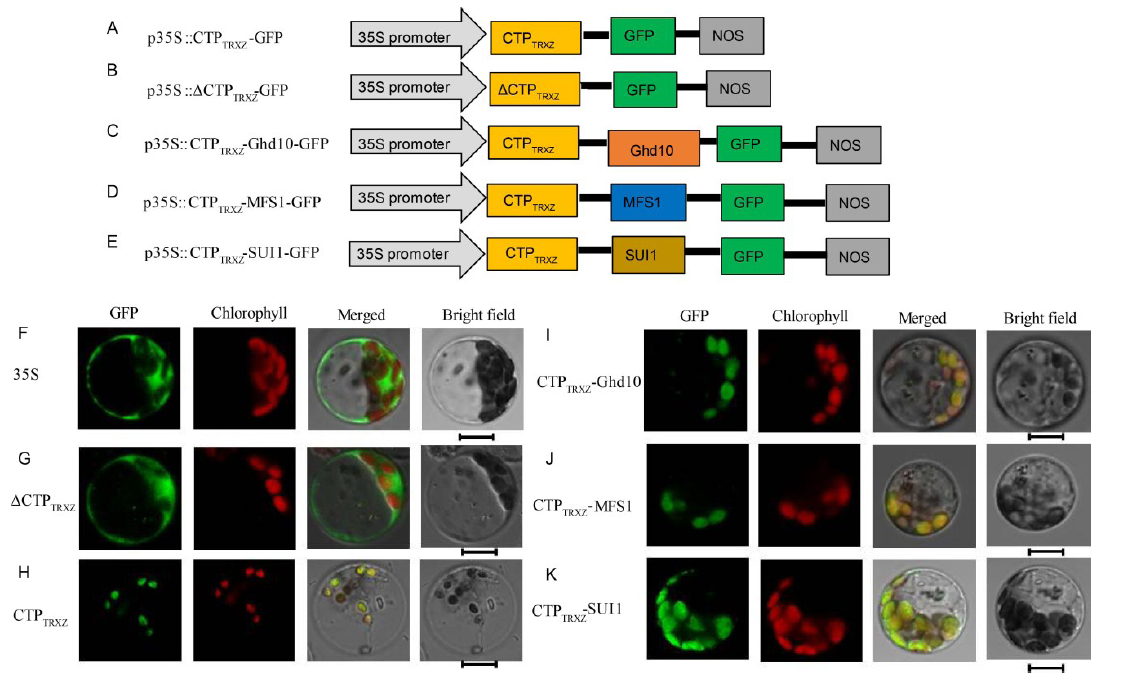
Fig. 2. Subcellular localization of TRXz and other proteins.A-E, Diagrams of vector construction; F, The free green fluorescent protein (GFP) protein as a control. G, The ΔCTPTRXz protein localized to the cytoplasm. H, The only N-terminal 56 amino acids of the TRXz and GFP fusion protein localized to chloroplast. I-K, The CTPTrxz-Ghd10, CTPTrxz-MFS1 and CTPTrxz-SUI1 fusion proteins localized to the chloroplasts. All of the GFP fusion proteins carried a C-terminal GFP tag. In F-K, GFP fluorescence, chlorophyll autofluorescence, merged GFP and chlorophyll fluorescence, and bright-field images are shown. Bar = 5 μm.
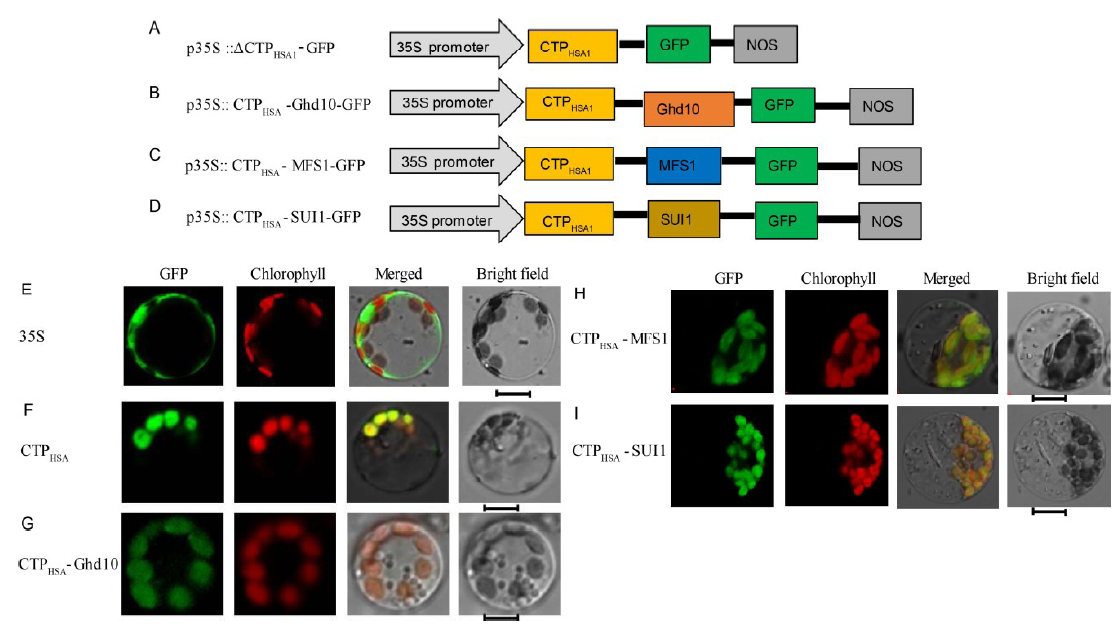
Fig. 3. Subcellular localization of HSA1 and other proteins.A-D, Diagrams of vector construction. E, The free green fluorescent protein (GFP) protein as a control. F, The only N-terminal 54 amino acids of the HSA1 and GFP fusion protein localized to chloroplast. G-I, The CTPHSA1-Ghd10, CTPHSA1-MFS1 and CTPHSA1-SUI1 fusion proteins localized to the chloroplasts. All of the GFP fusion proteins carried a C-terminal GFP tag. In E-I, GFP fluorescence, chlorophyll auto fluorescence, merged GFP and chlorophyll fluorescence, and bright-field images are shown. Bar = 5 μm.
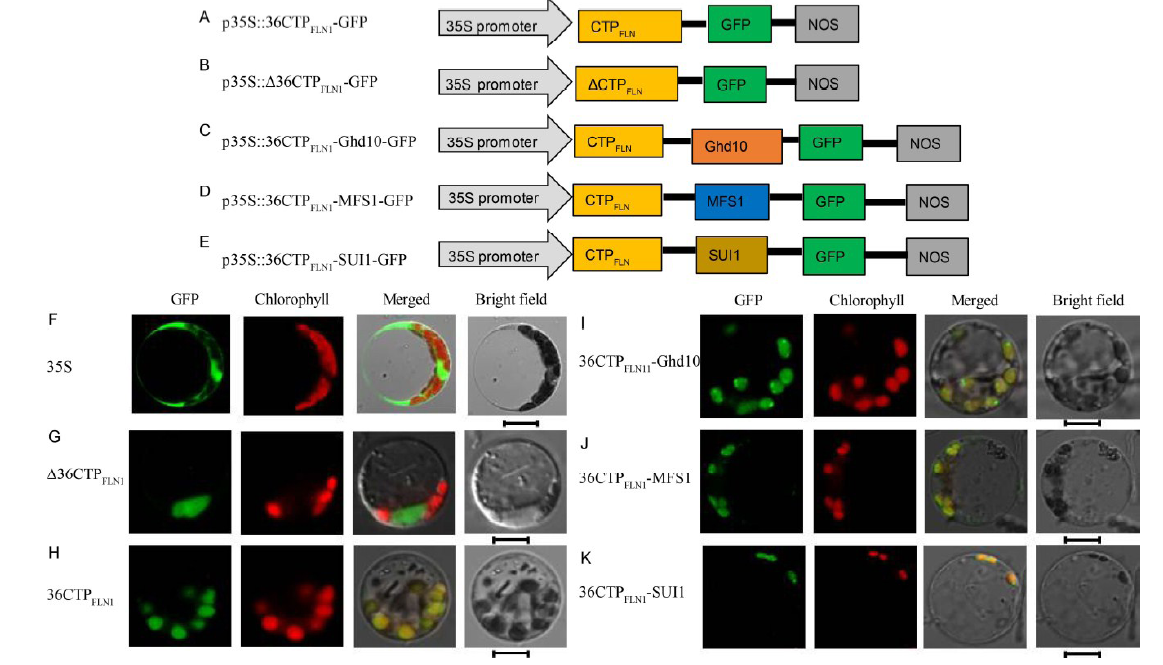
Fig. 4. Subcellular localization of FLN1 and other proteins. A-E, Diagrams of vector construction. F, The free green fluorescent protein (GFP) protein as a control. G, The Δ36CTPFLN1 protein localized to the cytoplasm. H, The only N-terminal 36 amino acids of the FLN1 and GFP fusion protein localized to chloroplast. I-K, The 36CTPFLN1-Ghd10, 36CTPFLN1-MFS1, and 36CTPFLN1-SUI1 fusion proteins localized to the chloroplasts. All of the GFP fusion proteins carried a C-terminal GFP tag. In F-K, GFP fluorescence, chlorophyll auto fluorescence, merged GFP and chlorophyll autofluorescence, and bright-field images are shown. Bar = 5 μm.
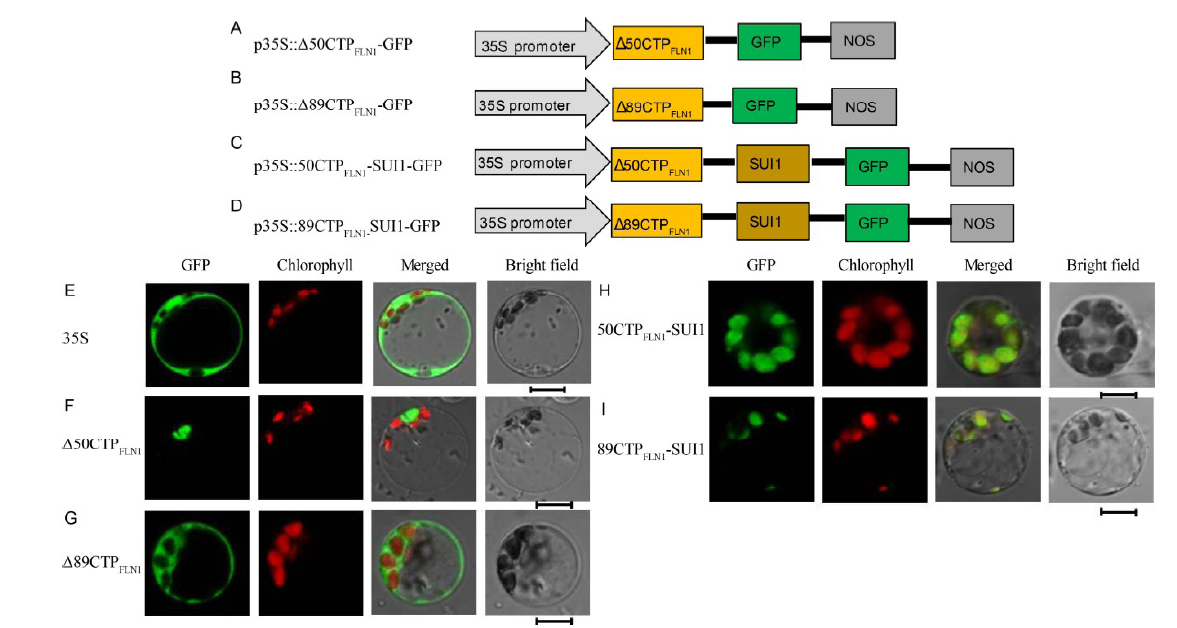
Fig. 5. Subcellular localization of other FLN1-related proteins. A-D, Diagrams of vector construction. E, The free green fluorescent protein (GFP) protein as a control. F, The Δ50CTPFLN1 protein localized to the cytoplasm. G, The Δ89CTPFLN1 protein localized to the cytoplasm. H and I, The 50CTPFLN1-SUI1 and 89CTPFLN1-SUI1 fusion proteins localized to the chloroplasts. All of the GFP fusion proteins carried a C-terminal GFP tag. In E-I, GFP fluorescence, chlorophyll autofluorescence, merged GFP and chlorophyll fluorescence, and bright-field images are shown. Bar = 5 μm.
| [1] | Fellerer C, Schweiger R, Schöngruber K, Soll J, Schwenkert S.2011. Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol Plant, 4(6): 1133-1145. |
| [2] | Hanke G, Mulo P.2013. Plant type ferredoxins and ferredoxin- dependent metabolism.Plant Cell Environ, 36(6): 1071-1084. |
| [3] | He L, Zhang S, Qiu Z N, Zhao J, Nie W D, Lin H Y, Zhu Z G, Zeng D L, Qian Q, Zhu L.2018. FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice.J Integr Plant Biol, 60(2): 94-111. |
| [4] | Hu S K, Dong G J, Xu J, Su Y, Shi Z Y, Ye W J, Li Y Y, Li G M, Zhang B, Hu J, Qian Q, Zeng D L, Guo L B.2013. A point mutation in the zinc finger motif of RID1/EHD2/OsID1 protein leads to outstanding yield-related traits in japonica rice variety Wuyunjing 7. Rice, 6(1): 24. |
| [5] | Ivey III R A, Bruce B D.2000. In vivo and in vitro interaction of DnaK and a chloroplast transit peptide. Cell Stress Chaper, 5(1): 62-71. |
| [6] | Keegstra K, Bauerle C.1988. Targeting of proteins into chloroplasts. Physiol Plantarum, 9(1): 15-19. |
| [7] | Kim S, Lee D S, Choi I S, Ahn S J, Kim Y H, Bae H J.2010. Arabidopsis thaliana Rubisco small subunit transit peptide increases the accumulation of Thermotoga maritima endoglucanase Cel5A in chloroplasts of transgenic tobacco plants. Transgenic Res, 19(3): 489-497. |
| [8] | Leister D.2003. Chloroplast research in the genomic age.Trends Genet, 19(1): 47-56. |
| [9] | Li F.2016. Transit peptide-mediated location of yeast acyl-δ9 desaturase in plastid leads to biosynthesis and accumulation of palmitoleic acid in tobacco leaf tissue. [Master thesis]. Jinzhong, China: Shanxi Agriculture University. (in Chinese with English abstract) |
| [10] | Li X, Lin Z M, Chen Z J, Wang C H, Liu X, Wang F.2013. Chloroplast targeting signal of a rice Rubisco activase gene enhances transgene expression.Fujian J Agric Sci, 28(2): 95-100. (in Chinese with English abstract) |
| [11] | Long S P, Zhu X G, Naidu S L, Ort D R.2006. Can improvement in photosynthesis increase crop yields?Plant Cell Environ, 29(3): 315-330. |
| [12] | Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D.2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA, 99(19): 12246-12251. |
| [13] | Murashige T, Skoog F.1962. A revised medium for rapid growth and bioassays with tobacco cultures.Physiol Plantarum, 15(3): 473-497. |
| [14] | Qbadou S, Becker T, Mirus O, Ivo T, Soll J, Schleiff E.2006. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64.Embo J, 25(9): 1836-1847. |
| [15] | Qiu Z N, Kang S J, He L, Zhao J, Zhang S, Hu J, Zeng D L, Zhang G H, Dong G J, Gao Z Y, Ren D Y, Chen G, Guo L B, Qian Q, Zhu L.2018. The newly identified heat-stress sensitive albino 1 gene affects chloroplast development in rice.Plant Sci, 267: 168-179. |
| [16] | Ren D Y, Li Y F, Zhao F M, Sang X C, Shi J Q, Wang N, Guo S, Ling Y H, Zhang C W, Yang Z L, He G H.2013. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol, 162(2): 872-884. |
| [17] | Shen B R, Zhu C H, Yao Z, Cui L L, Zhang J J, Yang C W, He Z H, Peng X X.2017. An optimized transit peptide for effective targeting of diverse foreign proteins into chloroplasts in rice.Sci Rep, 7: 46231. |
| [18] | Sun Q, Zybailov B, Majeran W, Friso G, Olinares P D B, van W K J.2009. PPDB, the plant proteomics database at Cornell.Nucl Acids Res, 37: 969-974. |
| [19] | Sun X W, Feng P Q, Xu X M, Guo H L, Ma J F, Chi W, Lin R C, Lu C M, Zhang L X.2011. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus.Nat Commun, 2(2): 477. |
| [20] | Timmis J N, Ayliffe M A, Huang C Y, Martin W.2004. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes.Nat Rev Genet, 5(2): 123-135. |
| [21] | Tu Z J, Zou G X, Huang L C, Chen L, Dai L P, Gao Y H, Leng Y J, Zhu L, Zhang G H, Hu J, Ren D Y, Gao Z Y, Dong G J, Chen G, Guo L B, Qian Q, Zeng D L.2017. Identification and fine mapping of pale green leaf PGL11 in rice. Chin J Rice Sci, 31(5): 489-499. (in Chinese with English abstract) |
| [22] | van den Broeck G, Timko M P, Kausch A P, Cashmore A R, van Montagu M, Herrera-Estrella L.1985. Targeting of a foreign protein to chloroplasts by fusion to the transit peptide from the small subunit of ribulose 1,5-bisphosphate carboxylase.Nature, 313: 358-363. |
| [23] | von Heijne G, Steppuhn J, Herrmann R G.1989. Domain structure of mitochondrial and chloroplast targeting peptides.Febs J, 180(3): 535-545. |
| [24] | Wong E Y, Hironaka C M, Fischhoff D A.1992. Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol, 20(1): 81-93. |
| [25] | Wu L W, Ren D Y, Hu S K, Li G M, Dong G J, Jiang L, Hu X M, Ye W J, Cui Y T, Zhu L, Hu J, Zhang G H, Gao Z Y, Zeng D L, Qian Q, Guo L B.2016. Down-regulation of a nicotinate phosphoribosyltransferase gene,OsNaPRT1, leads to withered leaf tips. Plant Physiol, 171(2): 1085-1098. |
| [26] | Zhang Q F.2007. Strategies for developing green super rice.Proc Natl Acad Sci USA, 104(42): 16402-16409. |
| [27] | Zhao H M, Song W B, Lai J S.2013. Cloning of sorghum bicolor chloroplast transit peptide (CTP) of 5-enolpyruvylshikimate- 3-phosphate synthase (EPSPS) and its functional validation in transgenic maize (Zea mays). J Agric Biotechnol, 21(9): 1009-1018. (in Chinese with English abstract) |
| [28] | Zhao J, Kang S J, Rao Y C, Qiu Z N, Jie X U, Jiang H U, Zhang G H, Zeng D L, Guo L B, Qian Q.2015. Subcellular localization analysis of phosphatidylserine synthase (OsSUI1) in tobacco mesophyll cells. Chin J Rice Sci, 29(4): 343-349. (in Chinese with English abstract) |
| [29] | Zhong H, Teymouri F, Chapman B, Maqbool S B, Sabzikar R, El-Maghraby Y, Dale B, Sticklen M B.2003. The pea (Pisum sativum L.) rbcS transit peptide directs the Alcaligenes eutrophus polyhydroxybutyrate enzymes into the maize(Zea mays L.) chloroplasts. Plant Sci, 165(3): 455-462. |
| [30] | Zhu L, Hu J, Zhu K M, Fang Y X, Gao Z Y, He Y H, Zhang G H, Guo L B, Zeng D L, Dong G J, Yan M X, Liu J, Qian Q.2011. Identification and characterization ofSHORTENED UPPERMOST INTERNODE 1, a gene negatively regulating uppermost internode elongation in rice. Plant Mol Biol, 77: 475-487. |
| [1] | Ayut Kongpun, Tonapha Pusadee, Pennapa Jaksomsak, Kawiporn Chinachanta, Patcharin Tuiwong, Phukjira Chan-In, Sawika Konsaeng, Wasu Pathom-Aree, Suchila Utasee, Benjamaporn Wangkaew, Chanakan Prom-U-Thai. Abiotic and Biotic Factors Controlling Grain Aroma along Value Chain of Fragrant Rice: A Review [J]. Rice Science, 2024, 31(2): 142-158. |
| [2] | Sujeevan Rajendran, Hyeonseo Park, Jiyoung Kim, Soon Ju Park, Dongjin Shin, Jong-Hee Lee, Young Hun Song, Nam-Chon Paek, Chul Min Kim. Methane Emission from Rice Fields: Necessity for Molecular Approach for Mitigation [J]. Rice Science, 2024, 31(2): 159-178. |
| [3] | Zhu Chengqi, Ye Yuxuan, Qiu Tian, Huang Yafan, Ying Jifeng, Shen Zhicheng. Drought-Tolerant Rice at Molecular Breeding Eras: An Emerging Reality [J]. Rice Science, 2024, 31(2): 179-189. |
| [4] | Wu Lijuan, Han Cong, Wang Huimei, He Yuchang, Lin Hai, Wang Lei, Chen Chen, E Zhiguo. OsbZIP53 Negatively Regulates Immunity Response by Involving in Reactive Oxygen Species and Salicylic Acid Metabolism in Rice [J]. Rice Science, 2024, 31(2): 190-202. |
| [5] | Xie Shuwei, Shi Huanbin, Wen Hui, Liu Zhiquan, Qiu Jiehua, Jiang Nan, Kou Yanjun. Carbon Catabolite Repressor UvCreA is Required for Development and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2024, 31(2): 203-214. |
| [6] | Zheng Shaoyan, Chen Junyu, Li Huatian, Liu Zhenlan, Li Jing, Zhuang Chuxiong. Analysis of RNA Recognition and Binding Characteristics of OsCPPR1 Protein in Rice [J]. Rice Science, 2024, 31(2): 215-225. |
| [7] | Liu Dan, Zhao Huibo, Wang Zi’an, Xu Jing, Liu Yiting, Wang Jiajia, Chen Minmin, Liu Xiong, Zhang Zhihai, Cen Jiangsu, Zhu Li, Hu Jiang, Ren Deyong, Gao Zhenyu, Dong Guojun, Zhang Qiang, Shen Lan, Li Qing, Qian Qian, Hu Songping, Zhang Guangheng. Leaf Morphology Genes SRL1 and RENL1 Co-Regulate Cellulose Synthesis and Affect Rice Drought Tolerance [J]. Rice Science, 2024, 31(1): 103-117. |
| [8] | Wei Huanhe, Geng Xiaoyu, Zhang Xiang, Zhu Wang, Zhang Xubin, Chen Yinglong, Huo Zhongyang, Zhou Guisheng, Meng Tianyao, Dai Qigen. Grain Yield, Biomass Accumulation, and Leaf Photosynthetic Characteristics of Rice under Combined Salinity-Drought Stress [J]. Rice Science, 2024, 31(1): 118-128. |
| [9] | Masoumeh Kordi, Naser Farrokhi, Martin I. Pech-Canul, Asadollah Ahmadikhah. Rice Husk at a Glance: From Agro-Industrial to Modern Applications [J]. Rice Science, 2024, 31(1): 14-32. |
| [10] | Tian Yu, Sun Jing, Li Jiaxin, Wang Aixia, Nie Mengzi, Gong Xue, Wang Lili, Liu Liya, Wang Fengzhong, Tong Litao. Effects of Milling Methods on Rice Flour Properties and Rice Product Quality: A Review [J]. Rice Science, 2024, 31(1): 33-46. |
| [11] | Norhashila Hashim, Maimunah Mohd Ali, Muhammad Razif Mahadi, Ahmad Fikri Abdullah, Aimrun Wayayok, Muhamad Saufi Mohd Kassim, Askiah Jamaluddin. Smart Farming for Sustainable Rice Production: An Insight into Application, Challenge, and Future Prospect [J]. Rice Science, 2024, 31(1): 47-61. |
| [12] | Gao Ningning, Ye Shuifeng, Zhang Yu, Zhou Liguo, Ma Xiaosong, Yu Hanxi, Li Tianfei, Han Jing, Liu Zaochang, Luo Lijun. A β-Carotene Ketolase Gene NfcrtO from Subaerial Cyanobacteria Confers Drought Tolerance in Rice [J]. Rice Science, 2024, 31(1): 62-76. |
| [13] | Li Qianlong, Feng Qi, Wang Heqin, Kang Yunhai, Zhang Conghe, Du Ming, Zhang Yunhu, Wang Hui, Chen Jinjie, Han Bin, Fang Yu, Wang Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 552-565. |
| [14] | Ji Dongling, Xiao Wenhui, Sun Zhiwei, Liu Lijun, Gu Junfei, Zhang Hao, Matthew Tom Harrison, Liu Ke, Wang Zhiqin, Wang Weilu. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 598-612. |
| [15] | Prathap V, Suresh Kumar, Nand Lal Meena, Chirag Maheshwari, Monika Dalal, Aruna Tyagi. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses [J]. Rice Science, 2023, 30(6): 613-631. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||