
Rice Science ›› 2025, Vol. 32 ›› Issue (6): 797-812.DOI: 10.1016/j.rsci.2025.10.003
• Reviews • Previous Articles Next Articles
Lishali Desingu1, R. L. Visakh2, R. P. Sah3, Uday Chand Jha4, R. V. Manju1, Swapna Alex5, Radha Beena1( )
)
Received:2025-05-25
Accepted:2025-08-10
Online:2025-11-28
Published:2025-12-04
Contact:
Radha Beena (Lishali Desingu, R. L. Visakh, R. P. Sah, Uday Chand Jha, R. V. Manju, Swapna Alex, Radha Beena. Genetic Regulation of Phytic Acid Biosynthesis in Rice: Pathways and Breeding Approaches for Low-Phytate Varieties[J]. Rice Science, 2025, 32(6): 797-812.
Add to citation manager EndNote|Ris|BibTeX
| Gene/Enzyme | Species | Function in phytic acid regulation | Reference |
|---|---|---|---|
| IPK1 (Inositol pentakisphosphate 2-kinase) | Arabidopsis thaliana, Phaseolus vulgaris, Glycine max, Triticum aestivum, Oryza sativa | Regulates the last step of phytic acid synthesis, converting InsP5 to InsP6 | Fileppi et al ( et al ( |
| IPK2 (Inositol polyphosphate kinase 2) | A. thaliana, P. vulgaris, O. sativa, G. max | Facilitates the conversion of lower inositol phosphates into higher forms in lipid-dependent phytic acid production | Xu et al ( |
| INO1 (myo-Inositol-3-phosphate synthase 1) | G. max, O. sativa | Encodes myo-inositol-3-phosphate synthase | Hegeman et al ( et al ( |
| MIPS (myo-Inositol-1-phosphate synthase) | Hordeum vulgare, Zea mays, P. vulgaris, Arachis hypogea, Cicer arietinum, Cajanus cajan, G. max, Lablab purpureus, Medicago truncatula, Pisum sativum, Trifolium pratense, Vigna unguiculata | Involved in transformation of myo- inositol-3-phosphate from glucose-6-phosphate | Larson and Raboy ( et al ( |
| MIK (myo-Inositol kinase) | Z. mays, P. vulgaris, O. sativa | Converts myo-inositol to inositol monophosphate, which initiates phytic acid biosynthesis pathway | Shi et al ( |
| MRP (ABC transporter) | O. sativa, A. thaliana, T. aestivum, Z. mays | Accountable for lpa1 mutation, affecting the regulation of phytic acid pathway | Garcia et al ( |
| LPA1 (Low phytic acid 1), LPA2 (Low phytic acid 2), LPA3 (Low phytic acid 3) | T. aestivum, O. sativa, Z. mays | Mutation leads to reduced phytic acid content in seeds | Venegas et al ( et al ( |
| ITPK (Inositol 1,3,4-trisphosphate 5/6-kinase) | G. max, P. vulgaris, A. thaliana, O. sativa | Encodes inositol tetrakisphosphate kinase, which catalyzes the phosphorylation of inositol pentakisphosphate (IP5) to inositol hexakisphosphate (IP6) | Stiles et al ( |
Table 1. Major genes involved in phytic acid biosynthesis.
| Gene/Enzyme | Species | Function in phytic acid regulation | Reference |
|---|---|---|---|
| IPK1 (Inositol pentakisphosphate 2-kinase) | Arabidopsis thaliana, Phaseolus vulgaris, Glycine max, Triticum aestivum, Oryza sativa | Regulates the last step of phytic acid synthesis, converting InsP5 to InsP6 | Fileppi et al ( et al ( |
| IPK2 (Inositol polyphosphate kinase 2) | A. thaliana, P. vulgaris, O. sativa, G. max | Facilitates the conversion of lower inositol phosphates into higher forms in lipid-dependent phytic acid production | Xu et al ( |
| INO1 (myo-Inositol-3-phosphate synthase 1) | G. max, O. sativa | Encodes myo-inositol-3-phosphate synthase | Hegeman et al ( et al ( |
| MIPS (myo-Inositol-1-phosphate synthase) | Hordeum vulgare, Zea mays, P. vulgaris, Arachis hypogea, Cicer arietinum, Cajanus cajan, G. max, Lablab purpureus, Medicago truncatula, Pisum sativum, Trifolium pratense, Vigna unguiculata | Involved in transformation of myo- inositol-3-phosphate from glucose-6-phosphate | Larson and Raboy ( et al ( |
| MIK (myo-Inositol kinase) | Z. mays, P. vulgaris, O. sativa | Converts myo-inositol to inositol monophosphate, which initiates phytic acid biosynthesis pathway | Shi et al ( |
| MRP (ABC transporter) | O. sativa, A. thaliana, T. aestivum, Z. mays | Accountable for lpa1 mutation, affecting the regulation of phytic acid pathway | Garcia et al ( |
| LPA1 (Low phytic acid 1), LPA2 (Low phytic acid 2), LPA3 (Low phytic acid 3) | T. aestivum, O. sativa, Z. mays | Mutation leads to reduced phytic acid content in seeds | Venegas et al ( et al ( |
| ITPK (Inositol 1,3,4-trisphosphate 5/6-kinase) | G. max, P. vulgaris, A. thaliana, O. sativa | Encodes inositol tetrakisphosphate kinase, which catalyzes the phosphorylation of inositol pentakisphosphate (IP5) to inositol hexakisphosphate (IP6) | Stiles et al ( |
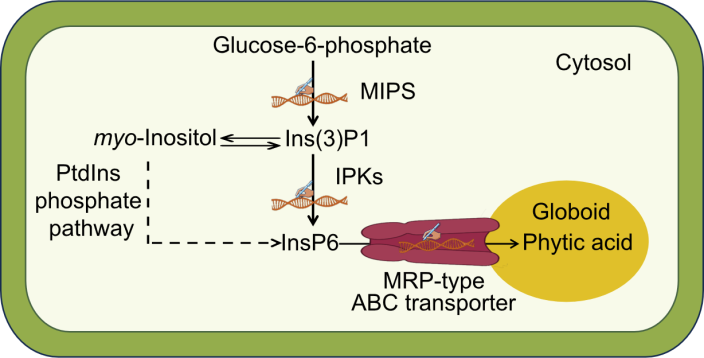
Fig. 2. Division of lpa mutations into three classes. PtdIns, Phosphatidylinositol; MIPS, myo-Inositol-3-phosphate synthase; IPK, Inositol polyphosphate kinase; Ins, myo-Inositol; MRP, Multidrug resistance-associated protein; ABC, ATP-binding cassette.
| Plant species | Targeted gene | Approach used | PA reduced (%) | Promoter employed | Yield penality | Reference |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | ITPK4, MRP5 | EMS Mu insertion | ‒ | ‒ | ‒ | Ren et al, |
| Triticum durum | TdMRP3 | TILLING | 85 | ‒ | 17.7% reduction of spikelet number per panicle and 34.5% reduction of grain number per spikelet | Frittelli et al, |
| Glycine max | GmIPK1 | CRISPR/Cas9 | < 25 | GmU6-10 promoter to drive sgRNAs and AtRPS5A promoter for Cas9 expression | ‒ | Song et al, |
| Triticum aestivum | TaIPK1 | CRISPR/Cas9 | 70 | TaU6 promoter to control sgRNAs and OsUbi promoter for Cas9 gene | ‒ | Ibrahim et al, |
| Zea mays | ZmMRP4 | EMS | < 36 | ‒ | ‒ | Abhijith et al, |
| Brassica napus | BnITPK1, BnITPK4 | CRISPR/Cas9 | < 35 | AtU6-26 promoter to drive expression of sgRNAs and Ubi4-2 promoter to drive Cas9 gene | No statistically significant reduction | Sashidhar et al, |
| Oryza sativa | OsITPK6 | CRISPR/Cas9 | < 32 | ‒ | Decreased grain yield and seed viability | Jiang et al, |
| Glycine max | GmMIPS1 | Antisense and RNAi | 38‒41 | Seed-specific vicilin promoter | Shorter plant height and root length, and less number of pods per plant | Kumar et al, |
| Glycine max | GmIPK2 | RNAi | 45 | Seed-specific vicilin promoter | No statistically significant reduction | Punjabi et al, |
| Triticum aestivum | TaABCC13 | RNAi | 22‒34 | pMCG161 RNAi vector | Grain filling and seed-setting rate were significantly affected | Bhati et al, |
| Oryza sativa | OsMIK | Hairpin RNA and artificial microRNA-mediated gene silencing | 14‒50 | Ole18 to drive seed-specific expression | ‒ | Li et al, |
| Oryza sativa | OsIPK1 | RNAi | 50-69 | Ole18 | ‒ | Ali et al, |
Table 2. Targeting genes to lower phytic acid (PA) levels in different plants.
| Plant species | Targeted gene | Approach used | PA reduced (%) | Promoter employed | Yield penality | Reference |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | ITPK4, MRP5 | EMS Mu insertion | ‒ | ‒ | ‒ | Ren et al, |
| Triticum durum | TdMRP3 | TILLING | 85 | ‒ | 17.7% reduction of spikelet number per panicle and 34.5% reduction of grain number per spikelet | Frittelli et al, |
| Glycine max | GmIPK1 | CRISPR/Cas9 | < 25 | GmU6-10 promoter to drive sgRNAs and AtRPS5A promoter for Cas9 expression | ‒ | Song et al, |
| Triticum aestivum | TaIPK1 | CRISPR/Cas9 | 70 | TaU6 promoter to control sgRNAs and OsUbi promoter for Cas9 gene | ‒ | Ibrahim et al, |
| Zea mays | ZmMRP4 | EMS | < 36 | ‒ | ‒ | Abhijith et al, |
| Brassica napus | BnITPK1, BnITPK4 | CRISPR/Cas9 | < 35 | AtU6-26 promoter to drive expression of sgRNAs and Ubi4-2 promoter to drive Cas9 gene | No statistically significant reduction | Sashidhar et al, |
| Oryza sativa | OsITPK6 | CRISPR/Cas9 | < 32 | ‒ | Decreased grain yield and seed viability | Jiang et al, |
| Glycine max | GmMIPS1 | Antisense and RNAi | 38‒41 | Seed-specific vicilin promoter | Shorter plant height and root length, and less number of pods per plant | Kumar et al, |
| Glycine max | GmIPK2 | RNAi | 45 | Seed-specific vicilin promoter | No statistically significant reduction | Punjabi et al, |
| Triticum aestivum | TaABCC13 | RNAi | 22‒34 | pMCG161 RNAi vector | Grain filling and seed-setting rate were significantly affected | Bhati et al, |
| Oryza sativa | OsMIK | Hairpin RNA and artificial microRNA-mediated gene silencing | 14‒50 | Ole18 to drive seed-specific expression | ‒ | Li et al, |
| Oryza sativa | OsIPK1 | RNAi | 50-69 | Ole18 | ‒ | Ali et al, |

Fig 3. Summary of phytic acid biosynthesis, storage and transport, nutritional impact, environmental and health aspects, and reduction strategies. MIPS, myo-Inositol-3-phosphate synthase; ITPK, Inositol 1,3,4-trisphosphate 5-/6-kinase; IPK, Inositol polyphosphate kinase; MRPS, Multidrug resistance-associated protein small subunit; ZIP, Zinc-regulated transporter like protein; YSL, Yellow stripe like; NRAMP, Natural resistance-associated macrophage protein transporter.
| [1] | Abhijith K P, Muthusamy V, Chhabra R, et al. 2020. Development and validation of breeder-friendly gene-based markers for lpa1-1 and lpa2-1 genes conferring low phytic acid in maize kernel. 3 Biotech, 10(3): 121. |
| [2] | Aggarwal S, Shukla V, Bhati K K, et al. 2015. Hormonal regulation and expression profiles of wheat genes involved during phytic acid biosynthesis pathway. Plants, 4(2): 298-319. |
| [3] | Aggarwal S, Kumar A, Bhati K K, et al. 2018. RNAi-mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases Fe and Zn accumulation. Front Plant Sci, 9: 259. |
| [4] | Ali N, Paul S, Gayen D, et al. 2013a. Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS One, 8(7): e68161. |
| [5] | Ali N, Paul S, Gayen D, et al. 2013b. RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice, 6(1): 12. |
| [6] | Andersen J R, Lübberstedt T. 2003. Functional markers in plants. Trends Plant Sci, 8: 554-560. |
| [7] | Bhati K K, Aggarwal S, Sharma S, et al. 2014. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci, 224: 74-85. |
| [8] | Bhati K K, Sharma S, Aggarwal S, et al. 2015. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front Plant Sci, 6: 488. |
| [9] | Bhati K K, Alok A, Kumar A, et al. 2016. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J Exp Bot, 67(14): 4379-4389. |
| [10] | Bhowmik A, Ojha D, Goswami D, et al. 2017. Inositol hexa phosphoric acid (phytic acid), a nutraceuticals, attenuates iron-induced oxidative stress and alleviates liver injury in iron overloaded mice. Biomed Pharmacother, 87: 443-450. |
| [11] | Boehm J D Jr. 2014. Molecular marker assisted backcross development and evaluation of an environmentally friendly, commercially acceptable low seed phytate soybean. Martin, Tennessee, USA: University of Tennessee. |
| [12] | Boehm J D Jr, Walker F R, Bhandari H S, et al. 2017. Seed inorganic phosphorus stability and agronomic performance of two low-phytate soybean lines evaluated across six southeastern US environments. Crop Sci, 57(5): 2555-2563. |
| [13] | Bohn L, Meyer A S, Rasmussen S K. 2008. Phytate: Impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B, 9(3): 165-191. |
| [14] | Campion B, Sparvoli F, Doria E, et al. 2009. Isolation and characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). Theor Appl Genet, 118(6): 1211-1221. |
| [15] | Campion B, Glahn R P, Tava A, et al. 2013. Genetic reduction of antinutrients in common bean (Phaseolus vulgaris L.) seed, increases nutrients and in vitro iron bioavailability without depressing main agronomic traits. Field Crops Res, 141: 27-37. |
| [16] | Cerino Badone F, Amelotti M, Cassani E, et al. 2012. Study of low phytic acid1-7 (lpa1-7), a new ZmMRP4 mutation in maize. J Hered, 103(4): 598-605. |
| [17] | Chang M X, Gu M, Xia Y W, et al. 2019. OsPHT1; 3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol, 179(2): 656-670. |
| [18] | Che J, Yamaji N, Miyaji T, et al. 2020. Node-localized transporters of phosphorus essential for seed development in rice. Plant Cell Physiol, 61(8): 1387-1398. |
| [19] | Chen H, Xiong L M. 2010. myo-Inositol-1-phosphate synthase is required for polar auxin transport and organ development. J Biol Chem, 285(31): 24238-24247. |
| [20] | Cominelli E, Confalonieri M, Carlessi M, et al. 2018. Phytic acid transport in Phaseolus vulgaris: A new low phytic acid mutant in the PvMRP1 gene and study of the PvMRPs promoters in two different plant systems. Plant Sci, 270: 1-12. |
| [21] | Cominelli E, Pilu R, Sparvoli F. 2020. Phytic acid and transporters: What can we learn from low phytic acid mutants. Plants, 9(1): 69. |
| [22] | Dong J S, Ma G J, Sui L Q, et al. 2019. Inositol pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol Plant, 12(11): 1463-1473. |
| [23] | Doria E, Galleschi L, Calucci L, et al. 2009. Phytic acid prevents oxidative stress in seeds: Evidence from a maize (Zea mays L.) low phytic acid mutant. J Exp Bot, 60(3): 967-978. |
| [24] | Du H, Liu L H, You L, et al. 2011. Characterization of an inositol 1, 3, 4-trisphosphate 5/6-kinase gene that is essential for drought and salt stress responses in rice. Plant Mol Biol, 77(6): 547-563. |
| [25] | Ertl D S, Young K A, Raboy V. 1998. Plant genetic approaches to phosphorus management in agricultural production. J Environ Qual, 27(2): 299-304. |
| [26] | Fileppi M, Galasso I, Tagliabue G, et al. 2010. Characterisation of structural genes involved in phytic acid biosynthesis in common bean (Phaseolus vulgaris L.). Mol Breed, 25(3): 453-470. |
| [27] | Frittelli A, Botticella E, Palombieri S, et al. 2023. The suppression of TdMRP3 genes reduces the phytic acid and increases the nutrient accumulation in durum wheat grain. Front Plant Sci, 14: 1079559. |
| [28] | Garcia O, Bouige P, Forestier C, et al. 2004. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J Mol Biol, 343(1): 249-265. |
| [29] | Gibson R S, Raboy V, King J C. 2018. Implications of phytate in plant-based foods for iron and zinc bioavailability, setting dietary requirements, and formulating programs and policies. Nutr Rev, 76(11): 793-804. |
| [30] | Gillaspy G E. 2013. The role of phosphoinositides and inositol phosphates in plant cell signaling. Adv Exp Med Biol, 991: 141-157. |
| [31] | Gillman J D, Baxter I, Bilyeu K. 2013. Phosphorus partitioning of soybean lines containing different mutant alleles of two soybean seed-specific adenosine triphosphate-binding cassette phytic acid transporter paralogs. Plant Genome, 6(1): 1-10. |
| [32] | Gupta R K, Gangoliya S S, Singh N K. 2015. Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J Food Sci Technol, 52(2): 676-684. |
| [33] | Gyani P C, Bollinedi H, Gopala Krishnan S, et al. 2020. Genetic analysis and molecular mapping of the quantitative trait loci governing low phytic acid content in a novel LPA rice mutant, PLM11. Plants, 9(12): 1728. |
| [34] | Hassler S, Lemke L, Jung B, et al. 2012. Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J, 72(5): 732-744. |
| [35] | Hegeman C E, Good L L, Grabau E A. 2001. Expression of d-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol, 125(4): 1941-1948. |
| [36] | Ibrahim S, Saleem B, Rehman N, et al. 2022. CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J Adv Res, 37: 33-41. |
| [37] | Ibrahim S, Andleeb T, Ramzan Khan M, et al. 2024. Genome-wide identification and characterization of Inositol Phosphokinase (IPK) gene family in wheat (Triticum aestivum L.). Plant Omics, 16(1): 1-10. |
| [38] | Jacob F, Hamid R, Ghorbanzadeh Z, et al. 2024. Genome-wide identification, characterization, and expression analysis of MIPS family genes in legume species. BMC Genomics, 25(1): 95. |
| [39] | Jiang M, Liu Y, Liu Y H, et al. 2019. Mutation of inositol 1,3,4-trisphosphate 5/6-kinase6 impairs plant growth and phytic acid synthesis in rice. Plants, 8(5): 114. |
| [40] | K R Y, Karjagi C G, Gangoliya S S, et al. 2025. Development of low phytate maize (Zea mays) inbred lines through marker-assisted pyramiding of lpa1 and lpa2 genes. Plant Mol Biol Rep, 43: 1262-1274. |
| [41] | Karmakar A, Bhattacharya S, Sengupta S, et al. 2020. RNAi-mediated silencing of ITPK gene reduces phytic acid content, alters transcripts of phytic acid biosynthetic genes, and modulates mineral distribution in rice seeds. Rice Sci, 27(4): 315-328. |
| [42] | Karner U, Peterbauer T, Raboy V, et al. 2004. myo-Inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J Exp Bot, 55: 1981-1987. |
| [43] | Kim S I, Tai T H. 2011. Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol Genet Genomics, 286(2): 119-133. |
| [44] | Kishor D S, Lee C, Lee D, et al. 2019. Novel allelic variant of Lpa1 gene associated with a significant reduction in seed phytic acid content in rice (Oryza sativa L.). PLoS One, 14(3): e0209636. |
| [45] | Kumar A, Kumar V, Krishnan V, et al. 2019. Seed targeted RNAi-mediated silencing of GmMIPS1 limits phytate accumulation and improves mineral bioavailability in soybean. Sci Rep, 9: 7744. |
| [46] | Kumar A, Sahu C, Panda P A, et al. 2020. Phytic acid content may affect starch digestibility and glycemic index value of rice (Oryza sativa L.). J Sci Food Agric, 100(4): 1598-1607. |
| [47] | Kumar A, Dash G K, Sahoo S K, et al. 2023. Phytic acid: A reservoir of phosphorus in seeds plays a dynamic role in plant and animal metabolism. Phytochem Rev, 22(5): 1281-1304. |
| [48] | Kumar V, Sinha A K, Makkar H P S, et al. 2010. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem, 120(4): 945-959. |
| [49] | Kumar V, Singh D, Sangwan P, et al. 2015. Management of environmental phosphorus pollution using phytases:Current challenges and future prospects. In: Kaushik G. Applied Environmental Biotechnology: Present Scenario and Future Trends. New Delhi, India: Springer: 97-114. |
| [50] | Kuo H F, Hsu Y Y, Lin W C, et al. 2018. Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J, 95(4): 613-630. |
| [51] | Kuwano M, Takaiwa F, Yoshida K T. 2009. Differential effects of a transgene to confer low phytic acid in caryopses located at different positions in rice panicles. Plant Cell Physiol, 50(7): 1387-1392. |
| [52] | Laha D, Parvin N, Hofer A, et al. 2019. Arabidopsis ITPK1 and ITPK2 have an evolutionarily conserved phytic acid kinase activity. ACS Chem Biol, 14(10): 2127-2133. |
| [53] | Larson S R, Raboy V. 1999. Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: Correspondence with a low phytic acid mutation. Theor Appl Genet, 99(1): 27-36. |
| [54] | Li W X, Huang J Z, Zhao H J, et al. 2014. Production of low phytic acid rice by hairpin RNA- and artificial microRNA-mediated silencing of OsMIK in seeds. Plant Cell Tissue Organ Cult, 119(1): 15-25. |
| [55] | Li Y T, Zhang J, Zhang X, et al. 2015. Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci, 230: 23-32. |
| [56] | Lin W X, Bai M Y, Peng C Y, et al. 2024. Genome editing toward biofortified soybean with minimal trade-off between low phytic acid and yield. aBiotech, 5(2): 196-201. |
| [57] | Liu Q L, Xu X H, Ren X L, et al. 2007. Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor Appl Genet, 114(5): 803-814. |
| [58] | Loewus F A, Murthy P P N. 2000. myo-Inositol metabolism in plants. Plant Sci, 150(1): 1-19. |
| [59] | Marathe A, Krishnan V, Vinutha T, et al. 2018. Exploring the role of inositol 1,3,4-trisphosphate 5/6 kinase-2 (GmITPK2) as a dehydration and salinity stress regulator in Glycine max (L.) Merr. through heterologous expression in E. coli. Plant Physiol Biochem, 123: 331-341. |
| [60] | Matsuno K, Fujimura T. 2014. Induction of phytic acid synthesis by abscisic acid in suspension-cultured cells of rice. Plant Sci, 217: 152-157. |
| [61] | Maurya K, Mani B, Singh B, et al. 2025. Editing cis-elements of OsPHO1;2 improved phosphate transport and yield in rice. Plant Biotechnol J, 23(9): 3864-3978.. |
| [62] | McCallum C M, Comai L, Greene E A, et al. 2000. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol, 123(2): 439-442. |
| [63] | Meis S J, Fehr W R, Schnebly S R. 2003. Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci, 43(4): 1336-1339. |
| [64] | Młodzińska E, Zboińska M. 2016. Phosphate uptake and allocation: A closer look at Arabidopsis thaliana L. and Oryza sativa L. Front Plant Sci, 7: 1198. |
| [65] | Muhammed A T P, Kumar A, Anilkumar C, et al. 2022. Understanding natural genetic variation for grain phytic acid content and functional marker development for phytic acid-related genes in rice. BMC Plant Biol, 22(1): 446. |
| [66] | Nunes A C S, Vianna G R, Cuneo F, et al. 2006. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta, 224(1): 125-132. |
| [67] | Nussaume L, Kanno S, Javot H, et al. 2011. Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci, 2: 83. |
| [68] | Panzeri D, Cassani E, Doria E, et al. 2011. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol, 191(1): 70-83. |
| [69] | Perera I, Seneweera S, Hirotsu N. 2018. Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice, 11(1): 4. |
| [70] | Perera I, Fukushima A, Akabane T, et al. 2019. Expression regulation of myo-inositol 3-phosphate synthase 1 (INO1) in determination of phytic acid accumulation in rice grain. Sci Rep, 9(1): 14866. |
| [71] | Pontoppidan K, Pettersson D, Sandberg A S. 2007. Peniophora lycii phytase is stabile and degrades phytate and solubilises minerals in vitro during simulation of gastrointestinal digestion in the pig. J Sci Food Agric, 87: 2700-2708. |
| [72] | Punjabi M, Bharadvaja N, Jolly M, et al. 2018. Development and evaluation of low phytic acid soybean by siRNA triggered seed specific silencing of Inositol polyphosphate 6-/3-/5-kinase gene. Front Plant Sci, 9: 804. |
| [73] | Qamar Z U, Hameed A, Ashraf M, et al. 2019. Development and molecular characterization of low phytate basmati rice through induced mutagenesis, hybridization, backcross, and marker assisted breeding. Front Plant Sci, 10: 1525. |
| [74] | Qin D, Toyonaga D, Saneoka H. 2021. Characterization of myo-inositol-1-phosphate synthase (MIPS) gene expression and phytic acid accumulation in oat (Avena sativa) during seed development. Cereal Res Commun, 50(3): 379-384. |
| [75] | Raboy V. 2003. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry, 64(6): 1033-1043. |
| [76] | Raboy V, 2009. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci, 177(4): 281-296. |
| [77] | Raboy V. 2020. Low phytic acid crops: Observations based on four decades of research. Plants, 9(2): 140. |
| [78] | Raboy V, Gerbasi P F, Young K A, et al. 2000. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1 Plant Physiol, 124(1): 355-368. |
| [79] | Raboy V, Young K A, Dorsch J A, et al. 2001. Genetics and breeding of seed phosphorus and phytic acid. J Plant Physiol, 158(4): 489-497. |
| [80] | Raboy V, Cichy K, Peterson K, et al. 2014. Barley (Hordeum vulgare L.) low phytic acid 1-1: An endosperm-specific, filial determinant of seed total phosphorus. J Hered, 105(5): 656-665. |
| [81] | Raboy V, Peterson K, Jackson C, et al. 2015. A substantial fraction of barley (Hordeum vulgare L.) low phytic acid mutations have little or no effect on yield across diverse production environments. Plants, 4(2): 225-239. |
| [82] | Rasmussen S K, Ingvardsen C R, Torp A M. 2010. Mutations in genes controlling the biosynthesis and accumulation of inositol phosphates in seeds. Biochem Soc Trans, 38(2): 689-694. |
| [83] | Ren Y Y, Jiang M D, Zhu J K, et al. 2024. Simultaneous mutations in ITPK4 and MRP5 genes result in a low phytic acid level without compromising salt tolerance in Arabidopsis. J Integr Plant Biol, 66(10): 2109-2125. |
| [84] | Sashidhar N, Harloff H J, Potgieter L, et al. 2020. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol J, 18(11): 2241-2250. |
| [85] | Sengupta S, Bhattacharya S, Karmakar A, et al. 2021. RNAi-mediated down-regulation of ITPK-2 enhanced inorganic phosphorus and minerals in the transgenic rice. J Biosci, 46: 32. |
| [86] | Shi J R, Wang H Y, Wu Y S, et al. 2003. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol, 131(2): 507-515. |
| [87] | Shi J R, Wang H Y, Hazebroek J, et al. 2005. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J, 42(5): 708-719. |
| [88] | Shukla V, Kaur M, Aggarwal S, et al. 2016. Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci Rep, 6: 39293. |
| [89] | Song J H, Shin G, Kim H J, et al. 2022. Mutation of GmIPK1 gene using CRISPR/Cas9 reduced phytic acid content in soybean seeds. Int J Mol Sci, 23(18): 10583. |
| [90] | Sparvoli F, Cominelli E. 2015. Seed biofortification and phytic acid reduction: A conflict of interest for the plant. Plants, 4(4): 728-755. |
| [91] | Stevenson-Paulik J, Bastidas R J, Chiou S T, et al. 2005. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci USA, 102: 12612-12617. |
| [92] | Stiles A R, Qian X, Shears S B, et al. 2008. Metabolic and signaling properties of an Itpk gene family in Glycine max. FEBS Lett, 582(13): 1853-1858. |
| [93] | Sun Y J, Thompson M, Lin G F, et al. 2007. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from maize: Molecular and biochemical characterization. Plant Physiol, 144(3): 1278-1291. |
| [94] | Sureshkumar S, Tamilkumar P, Thangavelu A U, et al. 2014. Marker-assisted introgression of lpa2 locus responsible for low-phytic acid trait into an elite tropical maize inbred (Zea mays L.). Plant Breed, 133(5): 566-578. |
| [95] | Suzuki M, Tanaka K, Kuwano M, et al. 2007. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): Implications for the phytic acid biosynthetic pathway. Gene, 405: 55-64. |
| [96] | Sylvia C, Sun J L, Zhang Y Q, et al. 2023. Genome-wide analysis of ATP binding cassette (ABC) transporters in peach (Prunus persica) and identification of a gene PpABCC1 involved in anthocyanin accumulation. Int J Mol Sci, 24(3): 1931. |
| [97] | Tagashira Y, Shimizu T, Miyamoto M, et al. 2015. Overexpression of a gene involved in phytic acid biosynthesis substantially increases phytic acid and total phosphorus in rice seeds. Plants, 4(2): 196-208. |
| [98] | Takagi D, Miyagi A, Tazoe Y, et al. 2020. Phosphorus toxicity disrupts Rubisco activation and reactive oxygen species defence systems by phytic acid accumulation in leaves. Plant Cell Environ, 43(9): 2033-2053. |
| [99] | Tang Y, Tan S T, Xue H W. 2013. Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase 2 is required for seed coat development. Acta Biochim Biophys Sin, 45(7): 549-560. |
| [100] | Teng W, Zhao Y Y, Zhao X Q, et al. 2017. Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat. Front Plant Sci, 8: 543. |
| [101] | Tp M A, Kumar A, Anilkumar C, et al. 2022. Understanding natural genetic variation for grain phytic acid content and functional marker development for phytic acid-related genes in rice. BMC Plant Biol, 22(1): 446. |
| [102] | Venegas J P, Graybosch R A, Wienhold B, et al. 2018. Biofortification of hard red winter wheat by genes conditioning low phytate and high grain protein concentration. Crop Sci, 58(5): 1942-1953. |
| [103] | Vincent J A, Stacey M, Stacey G, et al. 2015. Phytic acid and inorganic phosphate composition in soybean lines with independent IPK1 mutations. Plant Genome, 8(1): 1-10. |
| [104] | Wang F, Rose T, Jeong K, et al. 2016. The knowns and unknowns of phosphorus loading into grains, and implications for phosphorus efficiency in cropping systems. J Exp Bot, 67(5): 1221-1229. |
| [105] | Wang L N, Cui J, Zhang N, et al. 2024. OsIPK1 frameshift mutations disturb phosphorus homeostasis and impair starch synthesis during grain filling in rice. Plant Mol Biol, 114(5): 91. |
| [106] | Wild R, Gerasimaite R, Jung J Y, et al. 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science, 352: 986-990. |
| [107] | Xiang Y, Huang X Y, Zhao Y W, et al. 2024. Role of an ATP binding cassette (ABC) transporter MdABCI17 in the anthocyanin accumulation of apple. Sci Hortic, 323: 112502. |
| [108] | Xu J, Brearley C A, Lin W H, et al. 2005. A role of Arabidopsis inositol polyphosphate kinase, AtIPK2α, in pollen germination and root growth. Plant Physiol, 137(1): 94-103. |
| [109] | Yamaji N, Takemoto Y, Miyaji T, et al. 2017. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature, 541: 92-95. |
| [110] | Yang W, Liu X, Yu S, et al. 2024. The maize ATP-binding cassette (ABC) transporter ZmMRPA6 confers cold and salt stress tolerance in plants. Plant Cell Rep, 43(1): 13. |
| [111] | Yathish K R, Karjagi C G, Gangoliya S S, et al. 2022. Introgression of the low phytic acid locus (lpa2) into elite maize (Zea mays L.) inbreds through marker-assisted backcross breeding (MABB). Euphytica, 218(9): 127. |
| [112] | Yathish K R, Karjagi C G, Gangoliya S S, et al. 2023. Development of low-phytate maize inbred lines through marker-assisted introgression of lpa1. Crop Pasture Sci, 74(9): 843-855. |
| [113] | Ye H X, Zhang X Q, Broughton S, et al. 2011. A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct Integr Genomics, 11(1): 103-110. |
| [114] | Yuan F J, Zhu D H, Deng B, et al. 2009. Effects of two low phytic acid mutations on seed quality and nutritional traits in soybean (Glycine max L. Merr). J Agric Food Chem, 57(9): 3632-3638. |
| [115] | Zafar S A, Zaidi S S E A, Gaba Y, et al. 2020. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot, 71(2): 470-479. |
| [116] | Zhang F, Sun Y F, Pei W X, et al. 2015. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J, 82(4): 556-569. |
| [117] | Zhao H J, Liu Q L, Fu H W, et al. 2008. Effect of non-lethal low phytic acid mutations on grain yield and seed viability in rice. Field Crops Res, 108(3): 206-211. |
| [118] | Zhao H J, Cui H R, Xu X H, et al. 2013. Characterization of OsMIK in a rice mutant with reduced phytate content reveals an insertion of a rearranged retrotransposon. Theor Appl Genet, 126(12): 3009-3020. |
| [119] | Zhao H J, Frank T, Tan Y Y, et al. 2016. Disruption of OsSULTR3;3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol, 211(3): 926-939. |
| [120] | Zhou L J, Ye Y, Zhao Q, et al. 2018. Suppression of ROS generation mediated by higher InsP3 level is critical for the delay of seed germination in lpa rice. Plant Growth Regul, 85(3): 411-424. |
| [121] | Zhou L J, Asad M A U, Guan X Y, et al. 2024. Rice myo-inositol-3-phosphate synthase 2 (RINO2) alleviates heat injury-induced impairment in pollen germination and tube growth by modulating Ca2+ signaling and actin filament cytoskeleton. Plant J, 119(2): 861-878. |
| [122] | Zong G N, Shears S B, Wang H C. 2022. Structural and catalytic analyses of the InsP6 kinase activities of higher plant ITPKs. FASEB J, 36(7): e22380. |
| [123] | Zuber H, Davidian J C, Aubert G, et al. 2010. The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol, 154(2): 913-926. |
| [1] | Kunhikrishnan Hemalatha Dhanyalakshmi, Reshma Mohan, Sasmita Behera, Uday Chand Jha, Debashis Moharana, Ahalya Behera, Sini Thomas, Preman Rejitha Soumya, Rameswar Prasad Sah, Radha Beena. Next Generation Nutrition: Genomic and Molecular Breeding Innovations for Iron and Zinc Biofortification in Rice [J]. Rice Science, 2024, 31(5): 526-544. |
| [2] | Wu Lijuan, Han Cong, Wang Huimei, He Yuchang, Lin Hai, Wang Lei, Chen Chen, E Zhiguo. OsbZIP53 Negatively Regulates Immunity Response by Involving in Reactive Oxygen Species and Salicylic Acid Metabolism in Rice [J]. Rice Science, 2024, 31(2): 190-202. |
| [3] | Chen Yanhua, Wang Yaliang, Chen Huizhe, Xiang Jing, Zhang Yikai, Wang Zhigang, Zhu Defeng, Zhang Yuping. Brassinosteroids Mediate Endogenous Phytohormone Metabolism to Alleviate High Temperature Injury at Panicle Initiation Stage in Rice [J]. Rice Science, 2023, 30(1): 70-86. |
| [4] | Sheetal Bhadwal, Sucheta Sharma. Selenium Alleviates Carbohydrate Metabolism and Nutrient Composition in Arsenic Stressed Rice Plants [J]. Rice Science, 2022, 29(4): 385-396. |
| [5] | Wang Chenjiaozi, Zhao Mei, Shu Canwei, Zhou Erxun. Three Genes Related to Trehalose Metabolism Affect Sclerotial Development of Rhizoctonia solani AG-1 IA, Causal Agent of Rice Sheath Blight [J]. Rice Science, 2022, 29(3): 268-276. |
| [6] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [7] | Ramakrishna Wusirika, Kumari Anuradha, Rahman Nafeesa, Mandave Pallavi. Anticancer Activities of Plant Secondary Metabolites: Rice Callus Suspension Culture as a New Paradigm [J]. Rice Science, 2021, 28(1): 13-30. |
| [8] | Karmakar Aritra, Bhattacharya Sananda, Sengupta Shinjini, Ali Nusrat, Nath Sarkar Sailendra, Datta Karabi, K. Datta Swapan. RNAi-Mediated Silencing of ITPK Gene Reduces Phytic Acid Content, Alters Transcripts of Phytic Acid Biosynthetic Genes, and Modulates Mineral Distribution in Rice Seeds [J]. Rice Science, 2020, 27(4): 315-328. |
| [9] | Qiong Peng, Heping Han, Xia Yang, Lianyang Bai, Qin Yu, B. Powles Stephen. Quinclorac Resistance in Echinochloa crus-galli from China [J]. Rice Science, 2019, 26(5): 300-308. |
| [10] | Moe Kyi, Moh Moh Seinn, Zaw Htwe Aung, Kajihara Yoshinori, Yamakawa Takeo. Effects of Integrated Organic and Inorganic Fertilizers on Yield and Growth Parameters of Rice Varieties [J]. Rice Science, 2019, 26(5): 309-318. |
| [11] | Kalita Jyotirmay, Kumar Pradhan Amit, Moni Shandilya Zina, Tanti Bhaben. Arsenic Stress Responses and Tolerance in Rice: Physiological, Cellular and Molecular Approaches [J]. Rice Science, 2018, 25(5): 235-249. |
| [12] | Kaur Maninder, Asthir Bavita, Mahajan Gulshan. Variation in Antioxidants, Bioactive Compounds and Antioxidant Capacity in Germinated and Ungerminated Grains of Ten Rice Cultivars [J]. Rice Science, 2017, 24(6): 349-359. |
| [13] | Koteswara Reddy Chagam, Kimi Lalmuan, Haripriya Sundaramoorthy, Kang Nayoung. Effects of Polishing on Proximate Composition, Physico- Chemical Characteristics, Mineral Composition and Antioxidant Properties of Pigmented Rice [J]. Rice Science, 2017, 24(5): 241-252. |
| [14] | Mohibul Alam Khan Md, Haque Effi, Chandra Paul Narayan, Abdul Khaleque Md, M. S. Al-Garni Saleh, Rahman Mahfuzur, Tofazzal Islam Md. Enhancement of Growth and Grain Yield of Rice in Nutrient Deficient Soils by Rice Probiotic Bacteria [J]. Rice Science, 2017, 24(5): 264-273. |
| [15] | Kumar Verma Deepak, Prakash Srivastav Prem. Proximate Composition, Mineral Content and Fatty Acids Analyses of Aromatic and Non-Aromatic Indian Rice [J]. Rice Science, 2017, 24(1): 21-31. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||