
Rice Science ›› 2024, Vol. 31 ›› Issue (2): 215-225.DOI: 10.1016/j.rsci.2023.11.011
• Research Papers • Previous Articles Next Articles
Zheng Shaoyan,#( ), Chen Junyu#, Li Huatian, Liu Zhenlan, Li Jing, Zhuang Chuxiong(
), Chen Junyu#, Li Huatian, Liu Zhenlan, Li Jing, Zhuang Chuxiong( )
)
Received:2023-09-15
Accepted:2023-11-30
Online:2024-03-28
Published:2024-04-11
Contact:
Zhuang Chuxiong (About author:First author contact:#These authors contributed equally to this work
Zheng Shaoyan, Chen Junyu, Li Huatian, Liu Zhenlan, Li Jing, Zhuang Chuxiong. Analysis of RNA Recognition and Binding Characteristics of OsCPPR1 Protein in Rice[J]. Rice Science, 2024, 31(2): 215-225.
Add to citation manager EndNote|Ris|BibTeX
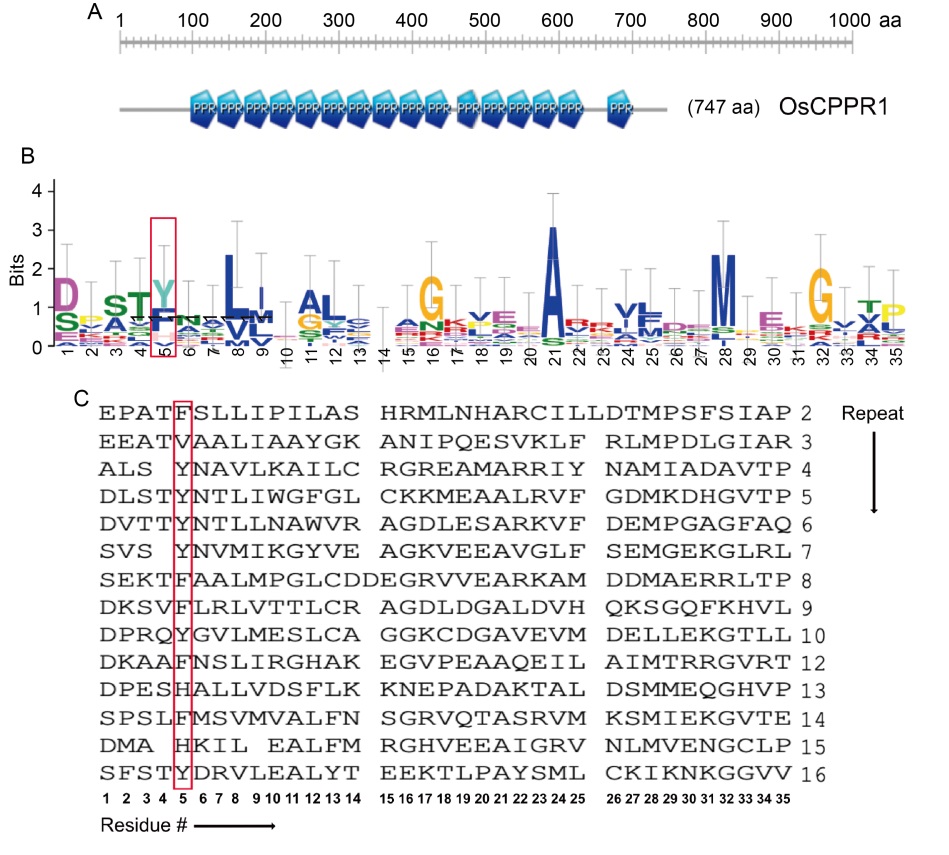
Fig. 1. Sequence alignment of pentatricopeptide repeat (PPR) protein in OsCPPR1. A, Schematic diagram of OsCPPR1 with its 16 PPR motifs. B, Sequence logo generated using PPR sequences of OsCPPR1 reveals relatively conserved amino acid positions within the PPR. C, Multiple sequence alignment of all 16 PPR motifs of OsCPPR1(aa 99-649). The determining residues for RNA recognition at the 5th positions are highlighted by a red box.
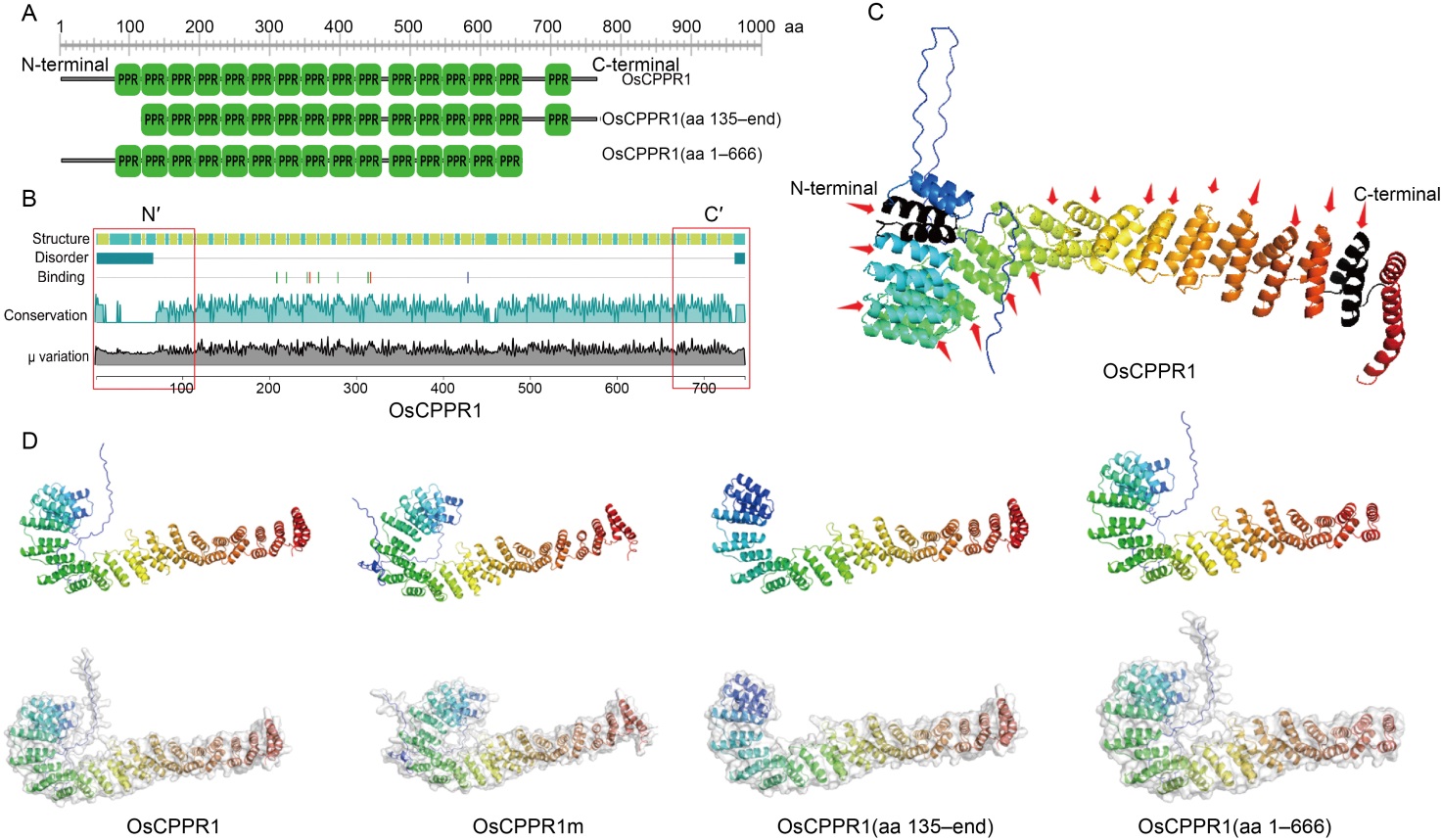
Fig. 2. Structure and similarity analysis of OsCPPR1, OsCPPR1(aa 135-end), and OsCPPR1(aa 1-666) proteins. A, Schematic diagram of OsCPPR1, OsCPPR1(aa 135-end), and OsCPPR1(aa 1-666). B, Secondary structure analyses of OsCPPR1, OsCPPR1(aa 135-end), and OsCPPR1(aa 1-666) were conducted using lamdba predict protein (λPP) (https://embed.predictprotein.org). The red box represents the corresponding missing pentatricopeptide repeat (PPR) motif region of OsCPPR1(aa 135-end) and OsCPPR1(aa 1-666). C, All 16 PPR motifs were showed in OsCPPR1 protein of the predicted three-dimensional structure. The PPR motifs were showed by the red arrows, and the black helix represented the PPR motif at the N- and C-terminal. D, Predicted three-dimensional structure (up) and the surface model structure (down) of different truncated OsCPPR1 proteins by Alphafold 2.
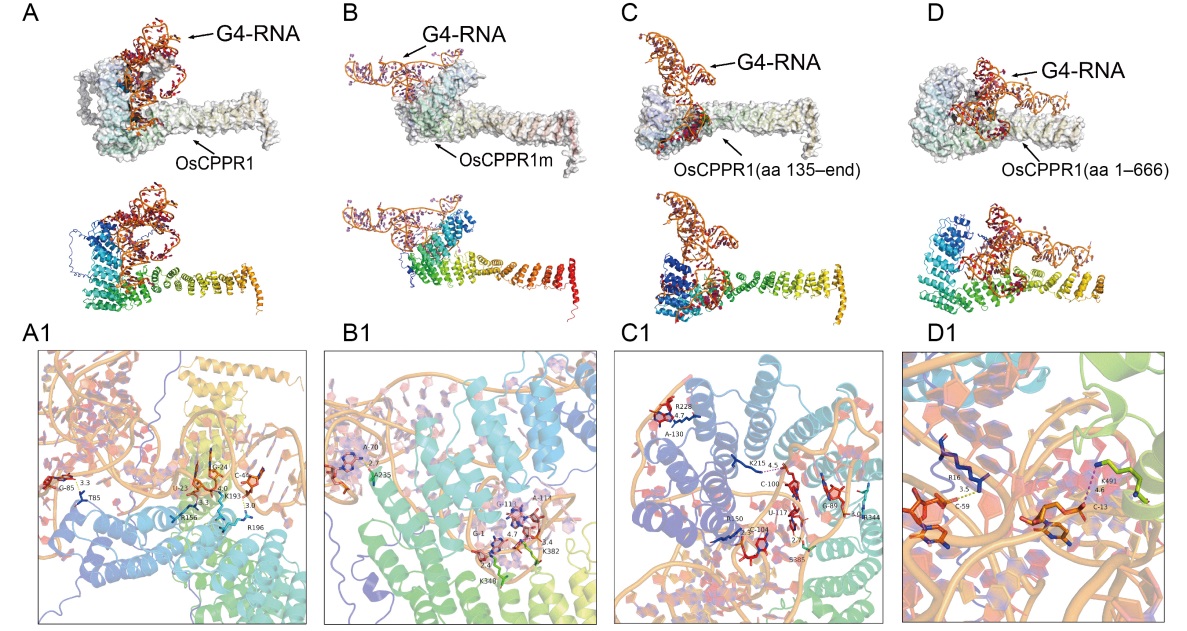
Fig. 4. Interaction analysis and prediction of OsGLK1-G4 RNA fragment with wild type and truncated OsCPPR1s. A-D, Base interaction prediction of OsGLK1-G4 RNA nucleotide with wild type (OsCPPR1, A) and truncated OsCPPR1s [OsCPPR1m, B; OsCPPR1(aa 135-end), C; OsCPPR1(aa 1-666), D] from the HDOCK server. A1-D1, The interaction analysis details between OsGLK1-G4 RNA nucleotide with wild type (OsCPPR1, A1) and truncated OsCPPR1s [OsCPPR1m, B1; OsCPPR1(aa 135-end), C1; OsCPPR1(aa 1-666), D1] from the HDOCK server.
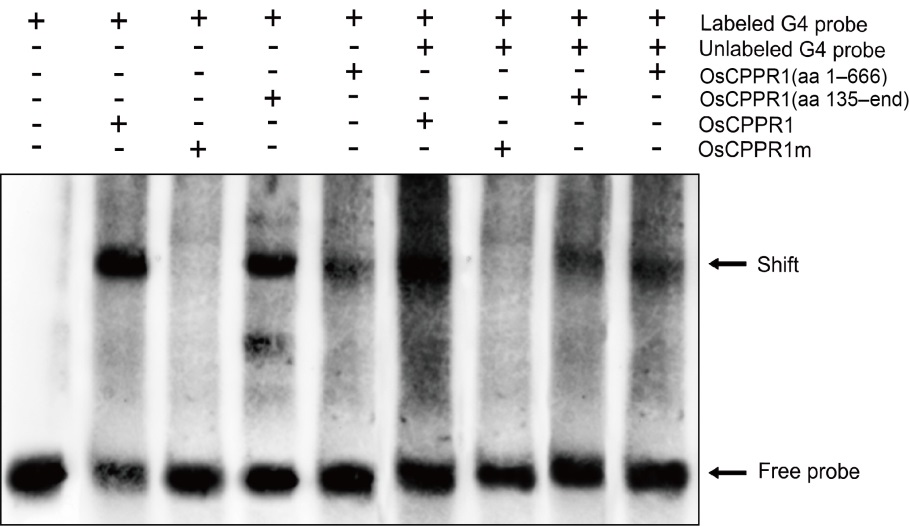
Fig. 5. RNA-electrophoretic mobility shift assay for binding of wild type and truncated OsCPPR1s to OsGLK1 G4-RNA fragment. RNA fragment G4 from the RIP-PCR (RNA immunoprecipitation quantitative polymerase chain reaction) analysis was end-labeled with biotin, and the same unlabeled RNA fragment (50-fold) was used as a cold competitor. The same concentration RNA fragment G4 was incubated with different truncated OsCPPR1 proteins, respectively.
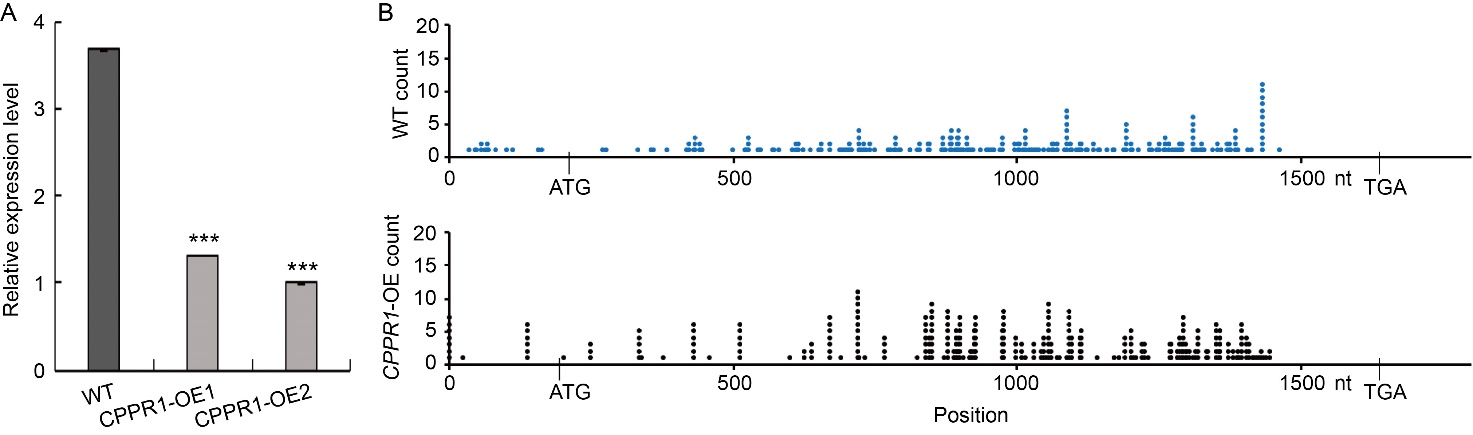
Fig. 6. Positions of 5′-RNA ligase-mediated rapid amplification of cDNA ends products on OsGLK1 in wild type (WT) and OsCPPR1 overexpression (CPPR1-OE) plants. A, qRT-PCR analysis of OsGLK1 in the flag levels of WT and CPPR1-OE (CPPR1-OE1 and CPPR1-OE2) plants at the flowering stage. Data are Mean ± SD (n = 3). ***, P < 0.001 according to the Student’s t-test. The ACTIN gene was used as an internal standard.B, Summary of cleavage sites on OsGLK1 transcripts in WT and CPPR1-OE (CPPR1-OE1 and CPPR1-OE2) (n > 100 clones).
| [1] | Abbas Y M, Pichlmair A, Górna M W, Superti-Furga G, Nagar B. 2013. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature, 494: 60-64. |
| [2] | Anandakrishnan R, Aguilar B, Onufriev A V. 2012. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res, 40: W537-W541. |
| [3] | Arenas-M A, González-Durán E, Gómez I, Burger M, Brennicke A, Takenaka M, Jordana X. 2018. The pentatricopeptide repeat protein MEF31 is required for editing at site 581 of the mitochondrial tatC transcript and indirectly influences editing at site 586 of the same transcript. Plant Cell Physiol, 59(2): 355-365. |
| [4] | Arnal N, Quadrado M, Simon M, Mireau H. 2014. A restorer-of- fertility like pentatricopeptide repeat gene directs ribonucleolytic processing within the coding sequence of rps3-rpl16 and orf240a mitochondrial transcripts in Arabidopsis thaliana. Plant J, 78(1): 134-145. |
| [5] | Ban T, Ke J Y, Chen R Z, Gu X, Tan M H, Zhou X E, Kang Y Y, Melcher K, Zhu J K, Xu H E. 2013. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J Biol Chem, 288(44): 31540-31548. |
| [6] | Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol, 65: 415-442. |
| [7] | Barkan A, Rojas M, Fujii S, Yap A, Chong Y S, Bond C S, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet, 8(8): e1002910. |
| [8] | Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol, 28(17): 5337-5347. |
| [9] | Biesiada M, Purzycka K J, Szachniuk M,. Blazewicz J, Adamiak R W. 2016. Automated RNA 3D structure prediction with RNAComposer. Methods Mol Biol, 1490: 199-215. |
| [10] | Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. 2011. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol, 155(4): 1678-1689. |
| [11] | Chen X Z, Feng F, Qi W W, Xu L M, Yao D S, Wang Q, Song R T. 2017. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant, 10(3): 427-441. |
| [12] | Cheng S F, Gutmann B, Zhong X, Ye Y T, Fisher M F, Bai F Q, Castleden I, Song Y, Song B, Huang J Y, Liu X, Xu X, Lim B L, Bond C S, Yiu S M, Small I. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J, 85(4): 532-547. |
| [13] | Dahan J, Mireau H. 2013. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol, 10(9): 1469-1476. |
| [14] | de Longevialle A F, Hendrickson L, Taylor N L, Delannoy E, Lurin C, Badger M, Millar A H, Small I. 2008. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J, 56(1): 157-168. |
| [15] | des Francs-Small C C, Sanglard L V P, Small I. 2018. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun Biol, 1: 166. |
| [16] | Ding Y H, Liu N Y, Tang Z S, Liu J, Yang W C. 2006. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell, 18(4): 815-830. |
| [17] | Gruber A R, Lorenz R, Bernhart S H, Neuböck R, Hofacker I L. 2008. The Vienna RNA websuite. Nucleic Acids Res, 36: W70-W74. |
| [18] | Gully B S, Cowieson N, Stanley W A, Shearston K, Small I D, Barkan A, Bond C S. 2015. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res, 43(3): 1918-1926. |
| [19] | Hall T M. 2016. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr Opin Struct Biol, 36: 116-121. |
| [20] | Hammani K, Okuda K, Tanz S K, Chateigner-Boutin A L, Shikanai T, Small I. 2009. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell, 21(11): 3686-3699. |
| [21] | Hammani K, Takenaka M, Miranda R, Barkan A. 2016. A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res, 44(9): 4278-4288. |
| [22] | Hao Y Y, Wang Y L, Wu M M, Zhu X P, Teng X, Sun Y L, Zhu J P, Zhang Y Y, Jing R N, Lei J, Li J F, Bao X H, Wang C M, Wang Y H, Wan J M. 2019. The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J Exp Bot, 70(18): 4705-4720. |
| [23] | Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. 2003. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J, 36( 4): 541-549. |
| [24] | Hattori M, Miyake H, Sugita M. 2007. A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J Biol Chem, 282(14): 10773-10782. |
| [25] | Huang W F, Zhang Y, Shen L Q, Fang Q, Liu Q, Gong C B, Zhang C, Zhou Y, Mao C, Zhu Y L, Zhang J H, Chen H P, Zhang Y, Lin Y J, Bock R, Zhou F. 2020. Accumulation of the RNA polymerase subunit RpoB depends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytol, 228(4): 1401-1416. |
| [26] | Jiang H C, Lu Q, Qiu S Q, Yu H H, Wang Z J, Yu Z C, Lu Y R, Wang L, Xia F, Wu Y Y, Li F, Zhang Q L, Liu G, Song D D, Ma C L, Ding Q, Zhang X B, Zhang L, Zhang X T, Li X, Zhang J W, Xiao J H, Li X H, Wang N Y, Ouyang Y D, Zhou F S, Zhang Q F. 2022. Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice. Proc Natl Acad Sci USA, 119(34): e2208759119. |
| [27] | Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern D B, Wollman F A, Vallon O. 2010. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell, 22(1): 234-248. |
| [28] | Kazama T, Toriyama K. 2003. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett, 544(1/3): 99-102. |
| [29] | Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. 2008. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J, 55(4): 619-628. |
| [30] | Ke J Y, Chen R Z, Ban T, Zhou X E, Gu X, Eileen Tan M H, Chen C, Kang Y Y, Brunzelle J S, Zhu J K, Melcher K, Xu H E. 2013. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol, 20(12): 1377-1382. |
| [31] | Koussevitzky S, Nott A, Mockler T C, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science, 316: 715-719. |
| [32] | Li X J, Zhang Y F, Hou M M, Sun F, Shen Y, Xiu Z H, Wang X M, Chen Z L, Sun S S M, Small I, Tan B C. 2014. Small kernel 1encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J, 79(5): 797-809. |
| [33] | Liu Y J, Xiu Z H, Meeley R, Tan B C. 2013. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell, 25(3): 868-883. |
| [34] | Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette M L, Mireau H, Peeters N, Renou J P, Szurek B, Taconnat L, Small I. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell, 16(8): 2089-2103. |
| [35] | Ma F, Hu Y C, Ju Y, Jiang Q R, Cheng Z J, Zhang Q,Sodmergen. 2017. A novel tetratricopeptide repeat protein, WHITE TO GREEN1, is required for early chloroplast development and affects RNA editing in chloroplasts. J Exp Bot, 68(21/22): 5829-5843. |
| [36] | Müller-McNicoll M, Rossbach O, Hui J Y, Medenbach J. 2019. Auto-regulatory feedback by RNA-binding proteins. J Mol Cell Biol, 11(10): 930-939. |
| [37] | O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz- Linneweber C, Sugita M, Small I. 2008. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol, 25(6): 1120-1128. |
| [38] | Pettersen E F, Goddard T D, Huang C C, Couch G S, Greenblatt D M, Meng E C, Ferrin T E. 2004. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem, 25(13): 1605-1612. |
| [39] | Pfalz J, Liere K, Kandlbinder A, Dietz K J, Oelmüller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell, 18(1): 176-197. |
| [40] | Prikryl J, Rojas M, Schuster G, Barkan A. 2011. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA, 108(1): 415-420. |
| [41] | Rovira A G, Smith A G. 2019. PPR proteins-orchestrators of organelle RNA metabolism. Physiol Plant, 166(1): 451-459. |
| [42] | Saha D, Prasad A M, Srinivasan R. 2007. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Biochem, 45(8): 521-534. |
| [43] | Schmitz-Linneweber C, Williams-Carrier R, Barkan A. 2005. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell, 17(10): 2791-2804. |
| [44] | Shen C C, Zhang D L, Guan Z Y, Liu Y X, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q X, Fan S L, Zou T T, Yin P. 2016. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat Commun, 7: 11285. |
| [45] | Small I D, Peeters N. 2000. The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci, 25(2): 45-47. |
| [46] | Sota F, Ian S. 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol, 191(1): 37-47. |
| [47] | Stern D B, Goldschmidt-Clermont M, Hanson M R. 2010. Chloroplast RNA metabolism. Annu Rev Plant Biol, 61: 125-155. |
| [48] | Tavares-Carreón F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingú-Vázquez M, Torres-Larios A, Pérez-Martínez X. 2008. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem, 283(3): 1472-1479. |
| [49] | Wang Z H, Zou Y J, Li X Y, Zhang Q Y, Chen L T, Wu H, Su D H, Chen Y L, Guo J X, Luo D, Long Y M, Zhong Y, Liu Y G. 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell, 18(3): 676-687. |
| [50] | Waters M T, Wang P, Korkaric M, Capper R G, Saunders N J, Langdale J A. 2009. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell, 21(4): 1109-1128. |
| [51] | Wu L L, Wu J, Liu Y X, Gong X D, Xu J L, Lin D Z, Dong Y J. 2016. The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast development under cold stress. Rice, 9(1): 67. |
| [52] | Xie T T, Chen D, Wu J, Huang X R, Wang Y F, Tang K L, Li J Y, Sun M X, Peng X B. 2016. Growing Slowly 1 locus encodes a PLS-type PPR protein required for RNA editing and plant development in Arabidopsis. J Exp Bot, 67(19): 5687-5698. |
| [53] | Yan Y M, Zhang D, Zhou P, Li B T, Huang S Y. 2017. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res, 45: W365-W373. |
| [54] | Yang H, Wang Y L, Tian Y L, Teng X, Lv Z H, Lei J, Duan E C, Dong H, Yang X, Zhang Y Y, Sun Y L, Chen X L, Bao X H, Chen R B, Gu C W, Zhang Y P, Jiang X K, Ma W Y, Zhang P C, Ji Y, Zhang Y, Wang Y H, Wan J M. 2023. Rice FLOURY ENDOSPERM22, encoding a pentatricopeptide repeat protein, is involved in both mitochondrial RNA splicing and editing and is crucial for endosperm development. J Integr Plant Biol, 65(3): 755-771. |
| [55] | Yang J Y, Zhang M, Wang X. 2018. Crystal structure of the chloroplast RNA editing factor MORF2. Biochem Biophys Res Commun, 495(2): 2038-2043. |
| [56] | Yap A, Kindgren P, Colas des Francs-Small C, Kazama T, Tanz S K, Toriyama K, Small I. 2015. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J, 81(5): 661-669. |
| [57] | Yin P, Li Q X, Yan C Y, Liu Y, Liu J J, Yu F, Wang Z, Long J F, He J H, Wang H W, Wang J W, Zhu J K, Shi Y G, Yan N E. 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature, 504: 168-171. |
| [58] | Zhang Y, Lu C M. 2019. The enigmatic roles of PPR-SMR proteins in plants. Adv Sci, 6(13): 1900361. |
| [59] | Zheng S Y, Li J, Ma L, Wang H L, Zhou H, Ni E D, Jiang D G, Liu Z L, Zhuang C X. 2019. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc Natl Acad Sci USA, 116(15): 7549-7558. |
| [60] | Zheng S Y, Dong J F, Lu J Q, Li J, Jiang D G, Yu H P, Ye S M, Bu W L, Liu Z L, Zhou H, Ding Y L, Zhuang C X. 2022. A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol, 234(5): 1678-1695. |
| [61] | Zhou W, Lu Q T, Li Q W, Wang L, Ding S H, Zhang A H, Wen X G, Zhang L X, Lu C M. 2017. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc Natl Acad Sci USA, 114(8): E1554-E1563. |
| [62] | Zoschke R, Kroeger T, Belcher S, Schöttler M A, Barkan A, Schmitz-Linneweber C. 2012. The pentatricopeptide repeat- SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J, 72(4): 547-558. |
| [63] | Zsigmond L, Szepesi Á, Tari I, Rigó G, Király A, Szabados L. 2012. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Sci, 182: 87-93. |
| [1] | Ayut Kongpun, Tonapha Pusadee, Pennapa Jaksomsak, Kawiporn Chinachanta, Patcharin Tuiwong, Phukjira Chan-In, Sawika Konsaeng, Wasu Pathom-Aree, Suchila Utasee, Benjamaporn Wangkaew, Chanakan Prom-U-Thai. Abiotic and Biotic Factors Controlling Grain Aroma along Value Chain of Fragrant Rice: A Review [J]. Rice Science, 2024, 31(2): 142-158. |
| [2] | Sujeevan Rajendran, Hyeonseo Park, Jiyoung Kim, Soon Ju Park, Dongjin Shin, Jong-Hee Lee, Young Hun Song, Nam-Chon Paek, Chul Min Kim. Methane Emission from Rice Fields: Necessity for Molecular Approach for Mitigation [J]. Rice Science, 2024, 31(2): 159-178. |
| [3] | Zhu Chengqi, Ye Yuxuan, Qiu Tian, Huang Yafan, Ying Jifeng, Shen Zhicheng. Drought-Tolerant Rice at Molecular Breeding Eras: An Emerging Reality [J]. Rice Science, 2024, 31(2): 179-189. |
| [4] | Wu Lijuan, Han Cong, Wang Huimei, He Yuchang, Lin Hai, Wang Lei, Chen Chen, E Zhiguo. OsbZIP53 Negatively Regulates Immunity Response by Involving in Reactive Oxygen Species and Salicylic Acid Metabolism in Rice [J]. Rice Science, 2024, 31(2): 190-202. |
| [5] | Xie Shuwei, Shi Huanbin, Wen Hui, Liu Zhiquan, Qiu Jiehua, Jiang Nan, Kou Yanjun. Carbon Catabolite Repressor UvCreA is Required for Development and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2024, 31(2): 203-214. |
| [6] | Liu Dan, Zhao Huibo, Wang Zi’an, Xu Jing, Liu Yiting, Wang Jiajia, Chen Minmin, Liu Xiong, Zhang Zhihai, Cen Jiangsu, Zhu Li, Hu Jiang, Ren Deyong, Gao Zhenyu, Dong Guojun, Zhang Qiang, Shen Lan, Li Qing, Qian Qian, Hu Songping, Zhang Guangheng. Leaf Morphology Genes SRL1 and RENL1 Co-Regulate Cellulose Synthesis and Affect Rice Drought Tolerance [J]. Rice Science, 2024, 31(1): 103-117. |
| [7] | Wei Huanhe, Geng Xiaoyu, Zhang Xiang, Zhu Wang, Zhang Xubin, Chen Yinglong, Huo Zhongyang, Zhou Guisheng, Meng Tianyao, Dai Qigen. Grain Yield, Biomass Accumulation, and Leaf Photosynthetic Characteristics of Rice under Combined Salinity-Drought Stress [J]. Rice Science, 2024, 31(1): 118-128. |
| [8] | Masoumeh Kordi, Naser Farrokhi, Martin I. Pech-Canul, Asadollah Ahmadikhah. Rice Husk at a Glance: From Agro-Industrial to Modern Applications [J]. Rice Science, 2024, 31(1): 14-32. |
| [9] | Tian Yu, Sun Jing, Li Jiaxin, Wang Aixia, Nie Mengzi, Gong Xue, Wang Lili, Liu Liya, Wang Fengzhong, Tong Litao. Effects of Milling Methods on Rice Flour Properties and Rice Product Quality: A Review [J]. Rice Science, 2024, 31(1): 33-46. |
| [10] | Norhashila Hashim, Maimunah Mohd Ali, Muhammad Razif Mahadi, Ahmad Fikri Abdullah, Aimrun Wayayok, Muhamad Saufi Mohd Kassim, Askiah Jamaluddin. Smart Farming for Sustainable Rice Production: An Insight into Application, Challenge, and Future Prospect [J]. Rice Science, 2024, 31(1): 47-61. |
| [11] | Gao Ningning, Ye Shuifeng, Zhang Yu, Zhou Liguo, Ma Xiaosong, Yu Hanxi, Li Tianfei, Han Jing, Liu Zaochang, Luo Lijun. A β-Carotene Ketolase Gene NfcrtO from Subaerial Cyanobacteria Confers Drought Tolerance in Rice [J]. Rice Science, 2024, 31(1): 62-76. |
| [12] | Li Qianlong, Feng Qi, Wang Heqin, Kang Yunhai, Zhang Conghe, Du Ming, Zhang Yunhu, Wang Hui, Chen Jinjie, Han Bin, Fang Yu, Wang Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 552-565. |
| [13] | Ji Dongling, Xiao Wenhui, Sun Zhiwei, Liu Lijun, Gu Junfei, Zhang Hao, Matthew Tom Harrison, Liu Ke, Wang Zhiqin, Wang Weilu. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 598-612. |
| [14] | Prathap V, Suresh Kumar, Nand Lal Meena, Chirag Maheshwari, Monika Dalal, Aruna Tyagi. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses [J]. Rice Science, 2023, 30(6): 613-631. |
| [15] | Serena Reggi, Elisabetta Onelli, Alessandra Moscatelli, Nadia Stroppa, Matteo Dell’Anno, Kiril Perfanov, Luciana Rossi. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Engineered Rice Lines [J]. Rice Science, 2023, 30(6): 587-597. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||