
Rice Science ›› 2020, Vol. 27 ›› Issue (4): 289-301.DOI: 10.1016/j.rsci.2020.05.005
• Research Paper • Previous Articles Next Articles
Shuting Yuan1,2,3,#, Chunjue Xu2,#, Wei Yan1,2, Zhenyi Chang1,2, Xingwang Deng2,3, Zhufeng Chen2( ), Jianxin Wu1(
), Jianxin Wu1( ), Xiaoyan Tang1,2(
), Xiaoyan Tang1,2( )
)
Received:2019-08-17
Accepted:2019-10-21
Online:2020-07-28
Published:2020-03-31
About author:#These authors contributed equally to this work
Shuting Yuan, Chunjue Xu, Wei Yan, Zhenyi Chang, Xingwang Deng, Zhufeng Chen, Jianxin Wu, Xiaoyan Tang. Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis[J]. Rice Science, 2020, 27(4): 289-301.
Add to citation manager EndNote|Ris|BibTeX
| Primer | Forward (5'-3') | Reverse('5'-3') |
|---|---|---|
| OsRAD1-P1 | CTCAACAGCCAAGCCGAA | AGGTGTGGAACGGTGGAA |
| OsRAD1-P2 | AGAGGGCGCGAAACCGTAGA | TCATACCACAATCCCTACGCATCAT |
| OsRAD1-P3 | AGAGGGCGCGAAACCGTAGA | AAAAACGGAGTCAAGCAATCATCAT |
| OsRAD1-P4 | TCCCCATCCTCTCCGTCGACTTCGT | ACTCAAGTACTCTGTGGAGGATGCCA |
| CR-1 target | CAAGGTCTACCTCAAGCGTG | |
| CR-2 target | CAGAACTTCCGTAATGTTGC | |
| CR-3 target | ACTGTTGCTTGAGCAGCAAA | |
| OsRAD1-P5 | GATCGAGGACCTTGAGTGGC | CACCTTGCTCCCACGATTCT |
| OsRAD1-P6 | GATGTTGGCGACCTCGTATT | CGCACAATCCCACTATCCTT |
| OsRAD1-P7 | GCCCAAATTGAATTGAAATCTGGCCTTTGAAGA | GCCCAAATTGAATTGAAATCTGGCCTTTGAAGA |
| OsRAD1-P8 | CATGAACAGATCTGCCCCGA | AGCCACAAAACCCCATCGAA |
| OsRAD1-P9 | CAGAACTTCCGTAATGTTGCTGG | TCGGTTTTGCCACTACCAGT |
| OsRAD1-P10 | GGGCCCAAAGCATTTTCCTT | TCCAAAAACGGAGTCAAGCA |
| OsRAD1-P11 | TGGTAGGGGTGGGATGCTTA | ACCACAATCCCTACGCATCA |
| SP-LR | GCGCGGTGTCATCTATGTTACT | CCCGACATAGATGCAATAACTTC |
| OsACTIN1-qPCR | GCTATGTACGTCGCCATCCA | GGACAGTGTGGCTGACACCAT |
| OsRAD1-qPCR | GATCGAGGACCTTGAGTGGC | AGGGGAACTCAATCTGCAAGT |
| OsRAD1.1-qPCR | AGAATCGTGGGAGCAAGGTG | AGCCCCTCCAGCAACATTAC |
| OsRAD1.2+3-qPCR | AGAATCGTGGGAGCAAGGTG | GGAAAATGCTTTGGGCCCTC |
| OsRAD1.2-qPCR | GTTGCAAGGCCAGGGTGTAA | GGTTCACCACGCTGTAGACT |
| OsRAD1.3-qPCR | CAAGGCCAGGTGTTCAAACG | GGTTCACCACGCTGTAGACT |
| MEL1-qPCR | TTACCCCCATTCCTATGAGCC | CAATCTTGCACACCTCCATAG |
| ZEP1-qPCR | CTGCCTCCAACATTAGTCAGC | CACTCGACCTAGAAGCTCCTG |
| PAIR1-qPCR | GGATGGACCCAGATTAACC | CTGTTTAGGTGCCACCCTGT |
| PAIR2-qPCR | TGCCAGAGGAGAGGACCATTC | CACGAGATGCTTGCTATTGAC |
| PAIR3-qPCR | GGAAGTTGAGCTGACGAACA | CAGTTCCCTGAGACAAGTTC |
| OsRAD1.1-BD | ATGAGCTCGTCGACGTCC | CTACGCATCATTTATCTCATA |
| OsRAD1.2-BD | ATGAGCTCGTCGACGTCC | TCACCACGCTGTAGACTGTT |
| OsRAD1.3-BD | ATGAGCTCGTCGACGTCC | TCACCACGCTGTAGACTGTT |
| OsRAD1M-BD | ATGAGCTCGTCGACGTCC | CTAAGCAATTCGGCTTGGCT |
| OsHUS1-AD | ATGAAGTTCAAGGCCTTCTT | TTAACTGCCAGGGTCAAGGA |
| OsRAD9-AD | ATGGAGCTGTCTATGAGCGG | CTAGTCCATGTAGTGCGGTG |
| OsRAD17-AD | ATGGGGAAGCGGCCGCCGGT | TCACCAATCTTCTATCTCAT |
Supplemental Table 1. Primers used in this study.
| Primer | Forward (5'-3') | Reverse('5'-3') |
|---|---|---|
| OsRAD1-P1 | CTCAACAGCCAAGCCGAA | AGGTGTGGAACGGTGGAA |
| OsRAD1-P2 | AGAGGGCGCGAAACCGTAGA | TCATACCACAATCCCTACGCATCAT |
| OsRAD1-P3 | AGAGGGCGCGAAACCGTAGA | AAAAACGGAGTCAAGCAATCATCAT |
| OsRAD1-P4 | TCCCCATCCTCTCCGTCGACTTCGT | ACTCAAGTACTCTGTGGAGGATGCCA |
| CR-1 target | CAAGGTCTACCTCAAGCGTG | |
| CR-2 target | CAGAACTTCCGTAATGTTGC | |
| CR-3 target | ACTGTTGCTTGAGCAGCAAA | |
| OsRAD1-P5 | GATCGAGGACCTTGAGTGGC | CACCTTGCTCCCACGATTCT |
| OsRAD1-P6 | GATGTTGGCGACCTCGTATT | CGCACAATCCCACTATCCTT |
| OsRAD1-P7 | GCCCAAATTGAATTGAAATCTGGCCTTTGAAGA | GCCCAAATTGAATTGAAATCTGGCCTTTGAAGA |
| OsRAD1-P8 | CATGAACAGATCTGCCCCGA | AGCCACAAAACCCCATCGAA |
| OsRAD1-P9 | CAGAACTTCCGTAATGTTGCTGG | TCGGTTTTGCCACTACCAGT |
| OsRAD1-P10 | GGGCCCAAAGCATTTTCCTT | TCCAAAAACGGAGTCAAGCA |
| OsRAD1-P11 | TGGTAGGGGTGGGATGCTTA | ACCACAATCCCTACGCATCA |
| SP-LR | GCGCGGTGTCATCTATGTTACT | CCCGACATAGATGCAATAACTTC |
| OsACTIN1-qPCR | GCTATGTACGTCGCCATCCA | GGACAGTGTGGCTGACACCAT |
| OsRAD1-qPCR | GATCGAGGACCTTGAGTGGC | AGGGGAACTCAATCTGCAAGT |
| OsRAD1.1-qPCR | AGAATCGTGGGAGCAAGGTG | AGCCCCTCCAGCAACATTAC |
| OsRAD1.2+3-qPCR | AGAATCGTGGGAGCAAGGTG | GGAAAATGCTTTGGGCCCTC |
| OsRAD1.2-qPCR | GTTGCAAGGCCAGGGTGTAA | GGTTCACCACGCTGTAGACT |
| OsRAD1.3-qPCR | CAAGGCCAGGTGTTCAAACG | GGTTCACCACGCTGTAGACT |
| MEL1-qPCR | TTACCCCCATTCCTATGAGCC | CAATCTTGCACACCTCCATAG |
| ZEP1-qPCR | CTGCCTCCAACATTAGTCAGC | CACTCGACCTAGAAGCTCCTG |
| PAIR1-qPCR | GGATGGACCCAGATTAACC | CTGTTTAGGTGCCACCCTGT |
| PAIR2-qPCR | TGCCAGAGGAGAGGACCATTC | CACGAGATGCTTGCTATTGAC |
| PAIR3-qPCR | GGAAGTTGAGCTGACGAACA | CAGTTCCCTGAGACAAGTTC |
| OsRAD1.1-BD | ATGAGCTCGTCGACGTCC | CTACGCATCATTTATCTCATA |
| OsRAD1.2-BD | ATGAGCTCGTCGACGTCC | TCACCACGCTGTAGACTGTT |
| OsRAD1.3-BD | ATGAGCTCGTCGACGTCC | TCACCACGCTGTAGACTGTT |
| OsRAD1M-BD | ATGAGCTCGTCGACGTCC | CTAAGCAATTCGGCTTGGCT |
| OsHUS1-AD | ATGAAGTTCAAGGCCTTCTT | TTAACTGCCAGGGTCAAGGA |
| OsRAD9-AD | ATGGAGCTGTCTATGAGCGG | CTAGTCCATGTAGTGCGGTG |
| OsRAD17-AD | ATGGGGAAGCGGCCGCCGGT | TCACCAATCTTCTATCTCAT |
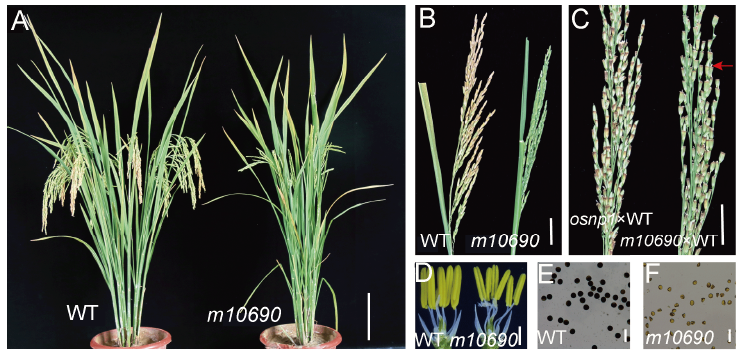
Fig. 1. Phenotypes of Huanghuazhan wild type (WT) and m10690 mutant plants.A, WT and m10690 mutant plants after heading. Scale bar, 10 cm. B, Seed-setting in WT and the mutant. Scale bar, 3 cm. C, Seed-setting of osnp1 and m10690 plants at 20 d after pollinated with the WT pollen. Red arrow indicates the seed-setting in the mutant. Scale bar, 2 cm. D, WT and m10690 spikelets with the palea and lemma removed to show the anthers. Scale bar, 1 mm. E, WT pollen grains stained with I2-KI. Scale bar, 100 μm. F, m10690 pollen grains stained with I2-KI. Scale bar, 100 μm.

Fig. 2. Transverse sections of anther in Huanghuazhan wild type (WT) and m10690 plants.WT anther sections are shown in A, C, E, G, I, K and M. The m10690 mutant anther sections are shown in B, D, F, H, J, L and N.E, Epidermis; En, Endothecium; M, Middle layer; T, Tapetal layer; PMC, Pollen mother cell; Dys, Dyads; Tds, Tetrads; MP, Mature pollen; Msp, Microspores. Scale bars, 20 μm.
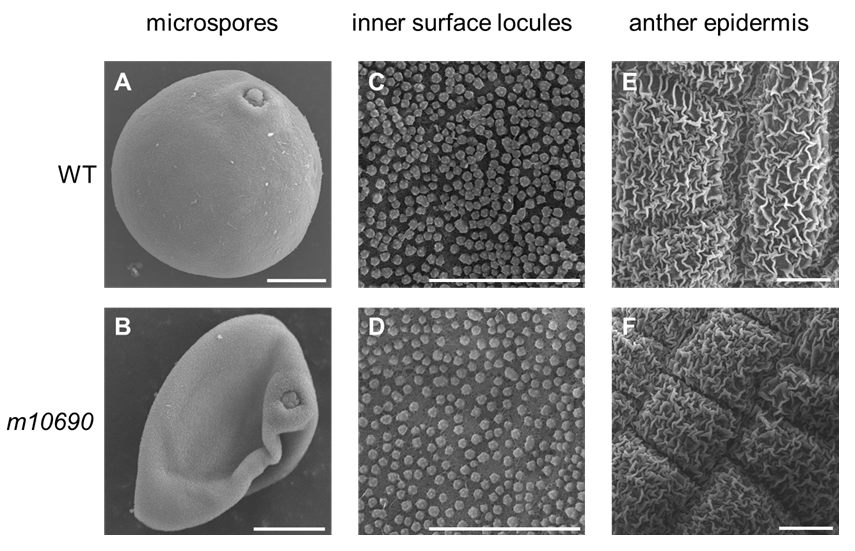
Supplemental Fig. 1. Scanning electron microscopy analysis of pollen grains, inner surface of tapetum, and outer surface of anther wall in the WT and m10690 mutant.Pollen grain of wild-type (WT) (A) and m10690 mutant (B), inner surface of WT tapetum (C) and m10690 mutant tapetum (D), and out surface of WT anther wall (E) and m10690 mutant anther wall (F) are shown. Scale=10μm.
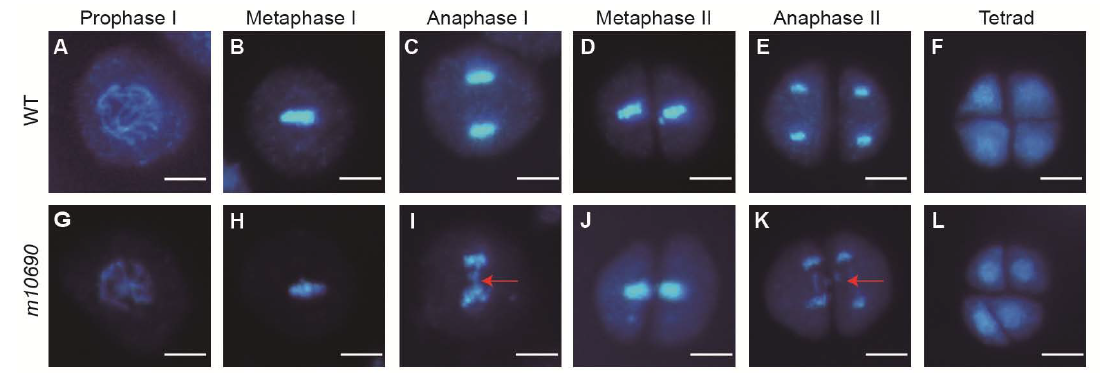
Fig. 3. Chromosome behavior during meiosis in pollen mother cells of Huanghuazhan wild type (WT) and m10690.A?F, Different meiosis stages for the WT. G?L, Different meiosis stages for the m10690 mutant. Red arrows indicate broken chromosome fragments. Scale bars, 10 μm.
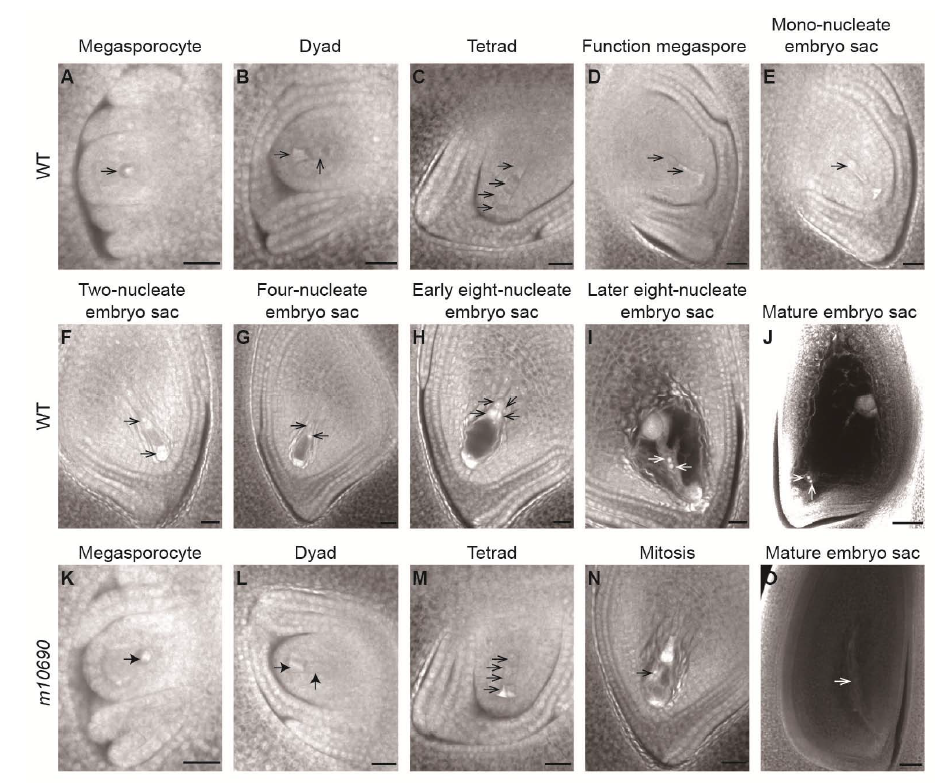
Fig. 4. Development of embryo sac in Huanghuazhan wild type (WT) and m10690 plants.A?J, The WT embryo sacs. K?O, The m10690 mutant embryo sacs. Arrows indicate the megaspores at various developmental stages. Scale bars, 20 μm.
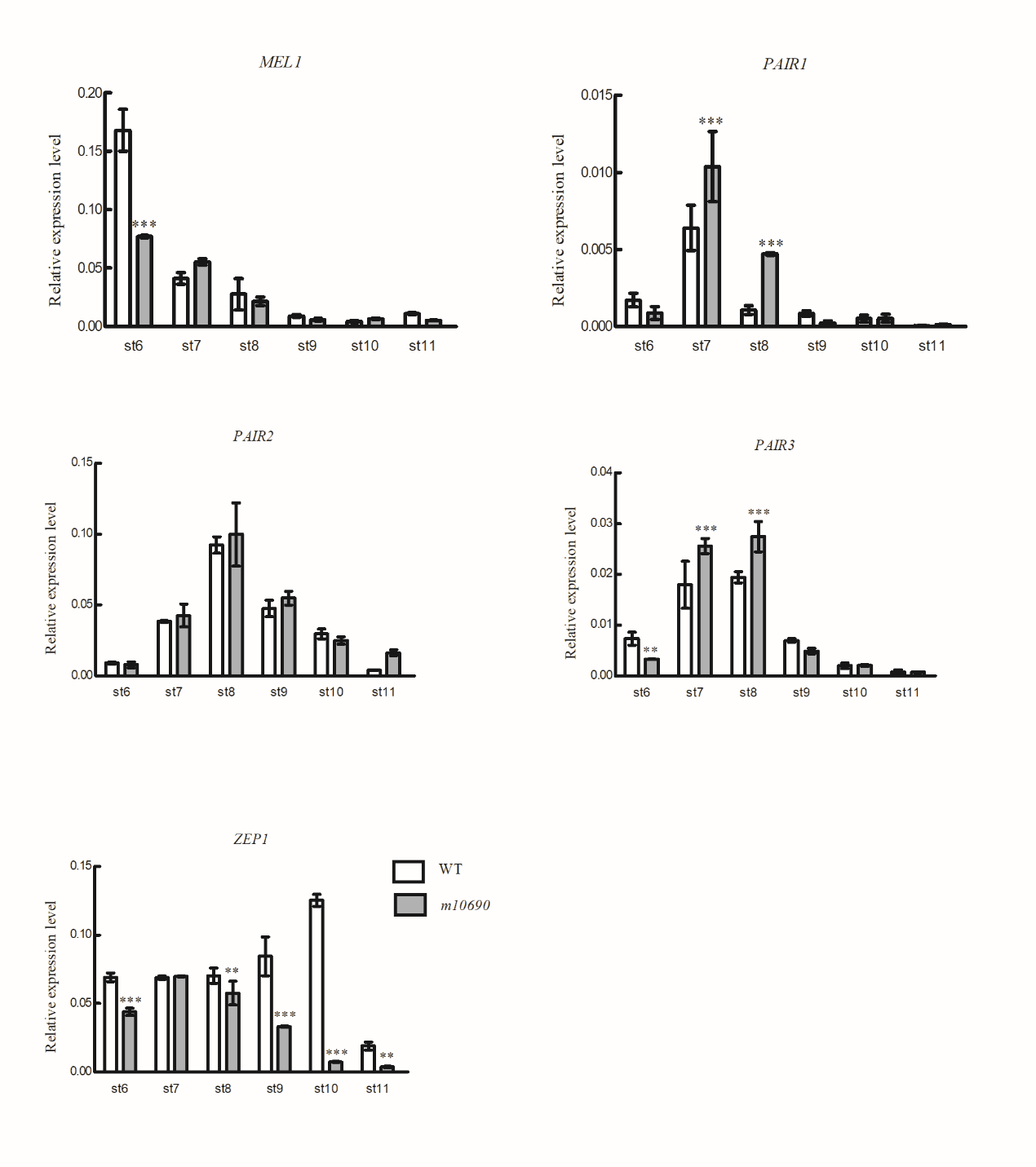
Supplemental Fig. 2. Expression of meiosis genes acting at prophase I in the WT and m10690 mutant anthers.Anthers were harvested from the WT and m10690 mutant plants at different developmental stages. Gene expression was determined using qRT-PCR. OsACTIN1 was used as internal control. Data are shown as means ± SD (n = 3). *** indicates significance at P?≤?0.001 and ** indicates significance at P?≤?0.01 by student’s t test.
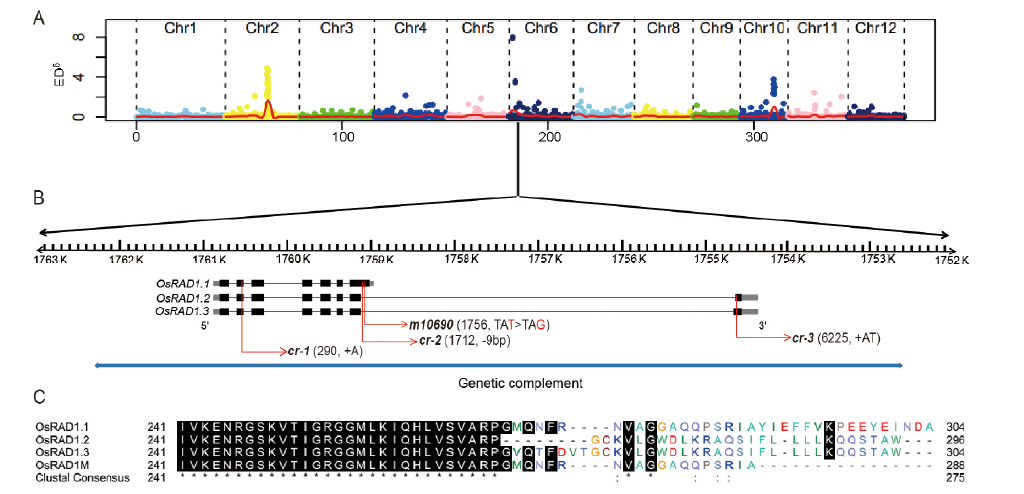
Fig. 5. Simultaneous identification of multiple causal mutations (SIMM) analysis of m10690 mutant and gene structure of OsRAD1.A, Mapping of the OsRAD1 gene based on the distribution of Euclidian Distance (ED6) values of SNPs along the rice chromosomes. B, OsRAD1 gene structure. The black boxes indicate exons, the gray boxes indicate 5′-UTR and 3′-UTR, the black lines indicate introns and the red arrows indicate the mutation sites in the original m10690 mutant and three other mutants generated by CRISPR knockout. The blue line represents the DNA fragment for transgenic complementation. C, Alignment of the C-terminal sequences of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1M.
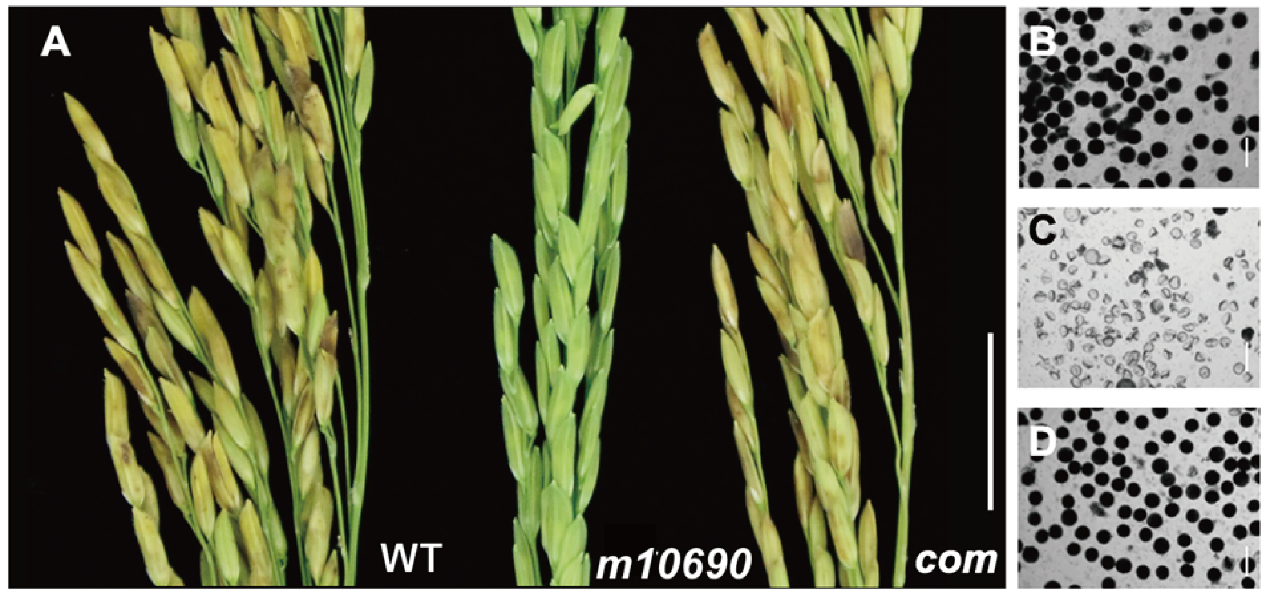
Supplemental Fig. 3. Transgenic complementation of the m10690 mutant.(A) Panicle of the WT HHZ, m10690 mutant, and transgenic-complemented m10690 plants. (B-D) I2-KI stained pollen grains of the WT (B), m10690 mutant (C), and transgenic-complemented osrad1 plants (D). Scale bars: A, 2cm; B-D, 100μm.
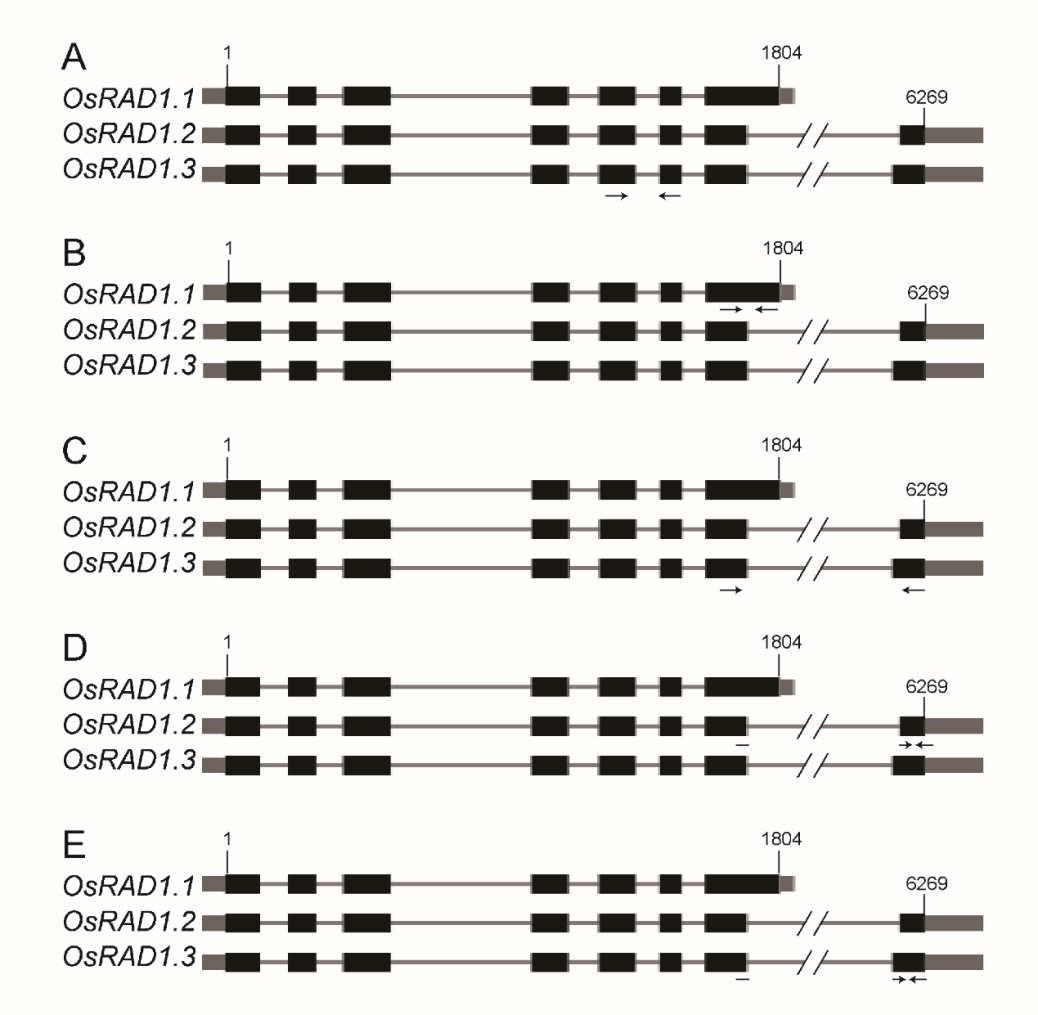
Supplemental Fig. 4. The positions of the primers for RT-PCR.(A), OsRAD1-qPCR; (B), OsRAD1.1-qPCR; (C), OsRAD1.2+3-qPCR; (D), OsRAD1.2-qPCR; (E), OsRAD1.3-qPCR. The black boxes indicate exons, the gray boxes indicate 5’UTR and 3’UTR, and the black lines indicate introns.
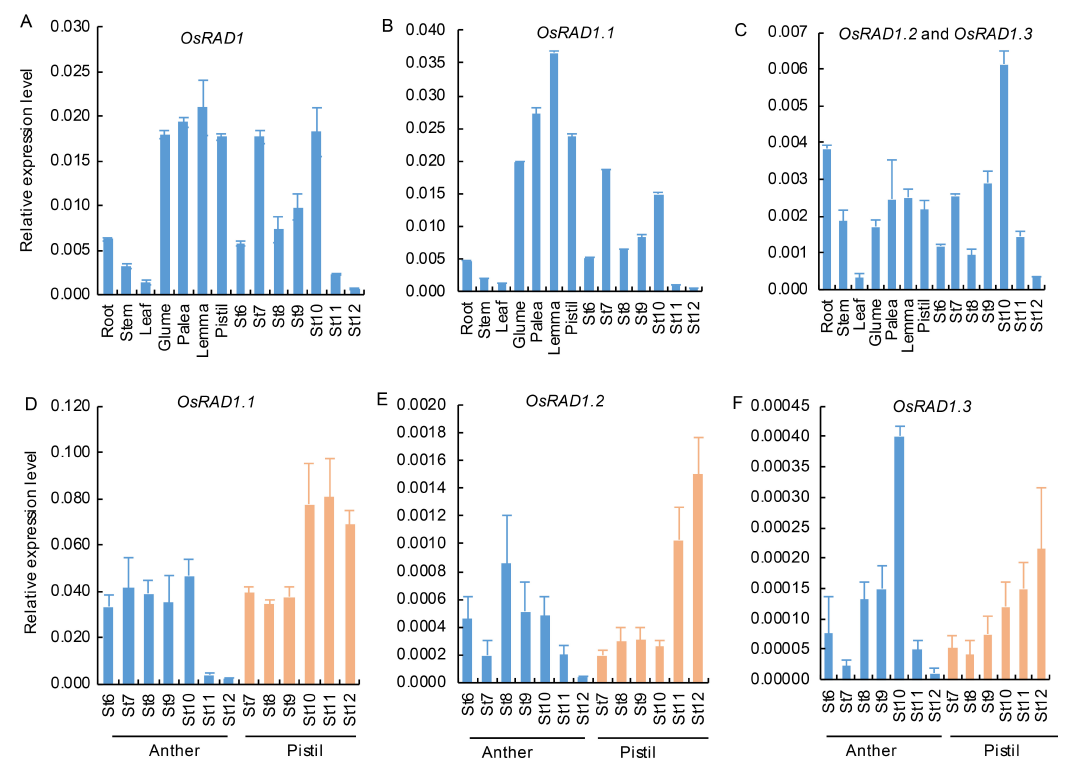
Fig. 6. Analysis of expression patterns of OsRAD1. A, Expression levels of OsRAD1 at different tissues and anther developmental stages. B, Expression levels of OsRAD1.1 at different tissues and anther developmental stages. C, Expression levels of OsRAD1.2 plus OsRAD1.3 at different tissues and anther developmental stages. D, Expression levels of OsRAD1.1 transcripts in anthers and pistils at different developmental stages. E, Expression levels of OsRAD1.2 transcripts in anthers and pistils at different developmental stages. F, Expression levels of OsRAD1.3 transcripts in anthers and pistils at different developmental stages.St6 to St12 are different developmental stages of anthers. Pistils and other tissues in A?C were harvested from plants at the flowering stage. Anthers and pistils in D?F were collected at St6 to St12. OsACTIN1 was used as the internal control. Data are Mean ± SD (n = 3).

Fig. 7. Phenotypes of CRISPR mutant alleles and transgenic plants overexpressing OsRAD1.2 plus OsRAD1.3.A, Wuyungeng 7. B, cr-1. C, cr-2. D, cr-3. E, cr-2 overexpressing OsRAD1.2. F, cr-2 overexpressing OsRAD1.3. G, Wuyungeng 7 over- expressing OsRAD1.2. H, Wuyungeng 7 overexpressing OsRAD1.3. The seed-setting and pollen grains stained with I2-KI are shown. White scale bars, 5 cm. Red scale bars, 100 μm. I, Relative expression levels of OsRAD1.2 and OsRAD1.3 in nontransgenic (N) and transgenic (P) plants of Wuyungeng 7 (WT) and cr-2 backgrounds. OsACTIN1 was used as the internal control. Data are shown as Mean ± SD (n = 3).
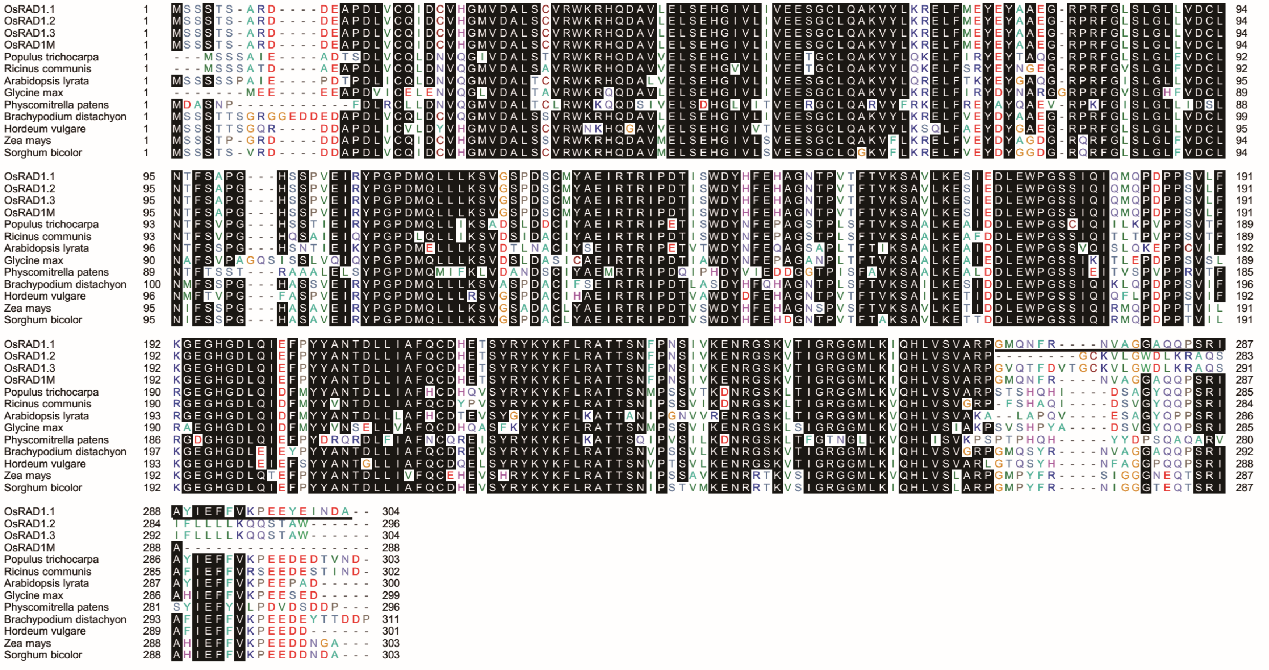
Supplemental Fig. 5. Alignment of OsRAD1.1, OsRAD1.2, OsRAD1.3, and OsRAD1.1M with RAD1 proteins from other plant species.Protein sequences were retrieved from NCBI by BLASTP search using OsRAD1.1 as query. The unique C-terminal domain sequence of OsRAD1.1 is underlined.
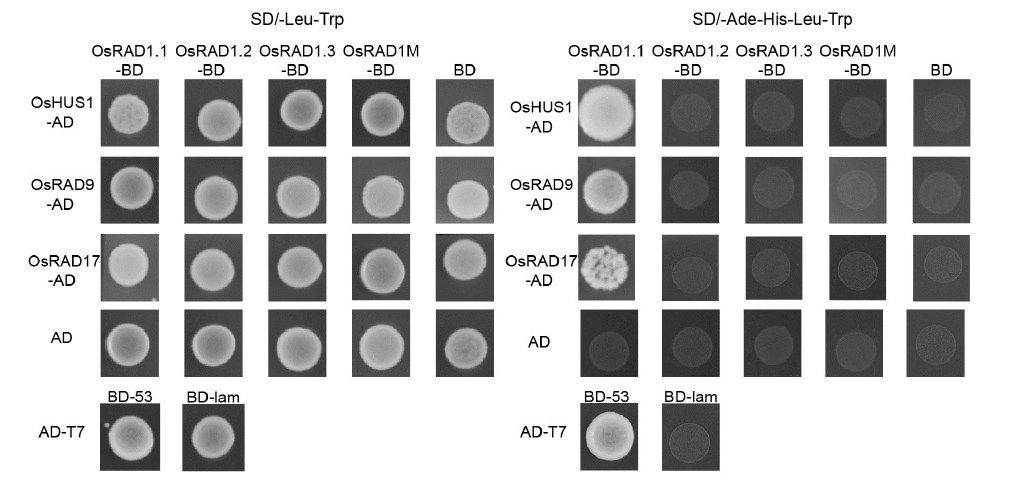
Fig. 8. Yeast two-hybrid assay of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1M interaction with RAD9, HUS1 and RAD17.SD/-Leu-Trp was used to test the co-transformation efficiency. The interactions were verified by the growth of yeast on selective medium SD/-Ade-His-Leu-Trp. The interaction of AD-T7 and BD-53 was included as the positive control, the interaction of AD-T7 and BD-lam was included as the negative control, and the interaction of BD and AD empty vectors was also included as the negative control.
| [1] | Bermudez V P, Lindsey-Boltz L A, Cesare A J, Maniwa Y, Griffith J D, Hurwitz J, Sancar A. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci USA, 100(4): 1633-1638. |
| [2] | Black D L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem, 72(1): 291-336. |
| [3] | Chang Z Y, Chen Z F, Wang N, Xie G, Lu J W, Yan W, Zhou J L, Tang X Y, Deng X W. 2016. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci USA, 113(49): 14145-14150. |
| [4] | Chaudhary S, Jabre I, Reddy A S N, Staiger D, Syed N H. 2019. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci, 24(6): 496-506. |
| [5] | Che L X, Wang K J, Tang D, Liu Q Q, Chen X J, Li Y F, Hu Q, Shen Y, Yu H X, Gu M H, Cheng Z K. 2014. OsHUS1 facilitates accurate meiotic recombination in rice. PLoS Genet, 10(6): e1004405. |
| [6] | Chen M, Manley J L. 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol, 10(11): 741-754. |
| [7] | Chen Z F, Lu J W, Lu Q Q, Wang N, Wang C X, Xie G, Zhou X Y, Tang X Y. 2014. Screening and analysis of male sterile mutants derived from elite indica cultivar Huanghuazhan. Guangdong Agric Sci, 41(19): 1-4. (in Chinese) |
| [8] | Doré A S, Kilkenny M L, Rzechorzek N J, Pearl L H. 2009. Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex-implications for clamp loading and regulation. Mol Cell, 34(6): 735-745. |
| [9] | Freire R, Murguía J R, Tarsounas M, Lowndes N F, Moens P B, Jackson S P. 1998. Human and mouse homologs of Schizosa- ccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: Linkage to checkpoint control and mammalian meiosis. Genes Dev, 12(16): 2560-2573. |
| [10] | Griffith J D, Lindsey-Boltz L A, Sancar A. 2002. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J Biol Chem, 277(18): 15233-15236. |
| [11] | Grushcow J M, Holzen T M, Park K J, Weinert T, Lichten M, Bishop D K. 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics, 153(2): 607-620. |
| [12] | Han L, Hu Z S, Liu Y H, Wang X Y, Hopkins K M, Lieberman H B, Hang H Y. 2010. Mouse Rad1 deletion enhances susceptibility for skin tumor development. Mol Cancer, 9(1): 67. |
| [13] | Heitzeberg F, Chen I P, Hartung F, Orel N, Angelis K J, Puchta H. 2004. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J, 38(6): 954-968. |
| [14] | Hu Q, Tang D, Wang H J, Shen Y, Chen X J, Ji J H, Du G J, Li Y F, Cheng Z K. 2016. The exonuclease homolog OsRAD1 promotes accurate meiotic double-strand break repair by suppressing nonhomologous end joining. Plant Physiol, 172(2): 1105-1116. |
| [15] | Hu Q, Zhang C, Xue Z H, Ma L J, Liu W, Shen Y, Ma B J, Cheng Z K. 2018. OsRAD17 is required for meiotic double-strand break repair and plays a redundant role with OsZIP4 in synaptonemal complex assembly. Front Plant Sci, 9: 1236. |
| [16] | Jaramillo-Lambert A, Harigaya Y, Vitt J, Villeneuve A, Engebrecht J. 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr Biol, 20(23): 2078-2089. |
| [17] | Kalsotra A, Cooper T A. 2011. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet, 12(10): 715-729. |
| [18] | Kelemen O, Convertini P, Zhang Z Y, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene, 514(1): 1-30. |
| [19] | Laloum T, Martín G, Duque P. 2018. Alternative splicing control of abiotic stress responses. Trends Plant Sci, 23(2): 140-150. |
| [20] | Lochlainn S Ó, Amoah S, Graham N S, Alamer K, Rios J J, Kurup S, Stoute A, Hammond J P, Østergaard L, King G J, White P J, Broadley M R. 2011. High resolution melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods, 7(1): 43. |
| [21] | Luo Q, Li Y F, Shen Y, Cheng Z K. 2014. Ten years of gene discovery for meiotic event control in rice. J Genet Genom, 41(3): 125-137. |
| [22] | Lyndaker A M, Lim P X, Mleczko J M, Diggins C E, Holloway J K, Holmes R J, Kan R, Schlafer D H, Freire R, Cohen P E, Weiss R S. 2013. Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet, 9(2): e1003320. |
| [23] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G. 2015. A robust CRISPR/Cas9 system for convenient, high- efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8(8): 1274-1284. |
| [24] | Melo J, Toczyski D. 2002. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol, 14(2): 237-245. |
| [25] | Naftelberg S, Schor I E, Ast G, Kornblihtt A R. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem, 84(1): 165-198. |
| [26] | Navadgi-Patil V M, Burgers P M. 2009. A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair, 8(9): 996-1003. |
| [27] | Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2004. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell, 16(4): 1008-1020. |
| [28] | Nonomura K, Nakano M, Eiquchi M, Suzuki T, Kurata N. 2006. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci, 119(2): 217-225. |
| [29] | Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2007. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell, 19(8): 2583-2594. |
| [30] | Ohashi E, Tsurimoto T. 2017. Functions of multiple clamp and clamp-loader complexes in eukaryotic DNA replication. Adv Exp Med Biol, 1042: 135-162. |
| [31] | Pan Q, Shai O, Lee L J, Frey B J, Blencowe B J. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet, 40(12): 1413-1415. |
| [32] | Peretz G, Arie L G, Bakhrat A, Abdu U. 2009. The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech Dev, 126: 677-686. |
| [33] | Shinohara M, Sakai K, Ogawa T, Shinohara A. 2003. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics, 164(3): 855-865. |
| [34] | Szakonyi D, Duque P. 2018. Alternative splicing as a regulator of early plant development. Front Plant Sci, 9: 1174. |
| [35] | Udell C M, Lee S K, Davey S. 1998. HRAD1 and MRAD1 encode mammalian homologues of the fission yeast rad1+ cell cycle checkpoint control gene. Nucl Acids Res, 26(17): 3971-3976. |
| [36] | Uri A, Martha K, Veronika B I, Anna B, Trudi S. 2007. An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J Cell Sci, 120(6): 1042-1049. |
| [37] | Vasileva A, Hopkins K M, Wang X Y, Weissbach M M, Friedman R A, Wolgemuth D J, Lieberman H B. 2013. The DNA damage checkpoint protein RAD9A is essential for male meiosis in the mouse. J Cell Sci, 126(17): 3927-3938. |
| [38] | Venclovas C, Thelen M P. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucl Acids Res, 28(13): 2481-2493. |
| [39] | Wang C L, Wang Y, Cheng Z J, Zhao Z G, Chen J, Sheng P K, Yu Y, Ma W W, Duan E C, Wu F Q, Liu L L, Qin R Z, Zhang X, Guo X P, Wang J L, Jiang L, Wan J M. 2016. The role of OsMSH4 in male and female gamete development in rice meiosis. J Exp Bot, 67(5): 1447-1459. |
| [40] | Wang M, Wang K J, Tang D, Wei C X, Li M, Shen Y, Chi Z C, Gu M H, Cheng Z K. 2010. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell, 22(2): 417-430. |
| [41] | Wang Y, Copenhaver G P. 2018. Meiotic recombination: Mixing it up in plants. Annu Rev Plant Biol, 69: 577-609. |
| [42] | Xu M, Bai L, Gong Y, Xie W, Hang H Y, Jiang T. 2009. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J Biol Chem, 284(31): 20457-20461. |
| [43] | Yan W, Chen Z F, Lu J W, Xu C J, Xie G, Li Y Q, Deng X W, He H, Tang X Y. 2017. Simultaneous identification of multiple causal mutations in rice. Front Plant Sci, 7: 2055. |
| [44] | Yuan W Y, Li X W, Chang Y X, Wen R Y, Chen G X, Zhang Q F, Wu C Y. 2009. Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J, 59(2): 303-315. |
| [45] | Zhang C B, Liu Y H, Hu Z S, An L L, He Y K, Hang H Y. 2011. Targeted deletion of mouse Rad1 leads to deficient cellular DNA damage responses. Protein Cell, 2(5): 410-422. |
| [46] | Zhang D B, Luo X, Zhu L. 2011. Cytological analysis and genetic control of rice anther development. J Genet Genom, 38(9): 379-390. |
| [47] | Zhou B B, Elledge S J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature, 408: 433-439. |
| [48] | Zou L, Cortez D, Elledge S J. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev, 16(2): 198-208. |
| [1] | Md. Dhin Islam, Adam H. Price, Paul D. Hallett. Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth [J]. Rice Science, 2023, 30(5): 459-472. |
| [2] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [3] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [4] | Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice [J]. Rice Science, 2022, 29(3): 237-246. |
| [5] | Vijayaraghavareddy Preethi, Xinyou Yin, C. Struik Paul, Makarla Udayakumar, Sreeman Sheshshayee. Responses of Lowland, Upland and Aerobic Rice Genotypes to Water Limitation During Different Phases [J]. Rice Science, 2020, 27(4): 345-354. |
| [6] | Hussain Kashif, Yingxing Zhang, Anley Workie, Riaz Aamir, Abbas Adil, Hasanuzzaman Rani Md., Hong Wang, Xihong Shen, Liyong Cao, Shihua Cheng. Association Mapping of Quantitative Trait Loci for Grain Size in Introgression Line Derived from Oryza rufipogon [J]. Rice Science, 2020, 27(3): 246-254. |
| [7] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [8] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| [9] | Fernando Polesi Luís, Bruder Silveira Sarmento Silene, Guidolin Canniatti-Brazaca Solange. Starch Digestibility and Functional Properties of Rice Starch Subjected to Gamma Radiation [J]. Rice Science, 2018, 25(1): 42-51. |
| [10] | Singh Bhupinder, Raja Reddy Kambham, Diaz Redoña Edilberto, Walker Timothy. Screening of Rice Cultivars for Morpho-Physiological Responses to Early-Season Soil Moisture Stress [J]. Rice Science, 2017, 24(6): 322-335. |
| [11] | Jini D., Joseph B.. Physiological Mechanism of Salicylic Acid for Alleviation of Salt Stress in Rice [J]. Rice Science, 2017, 24(2): 97-108. |
| [12] | Fernando Polesi Luís, Divino da Matta Junior Manoel, Bruder Silveira Sarmento Silene, Guidolin Canniatti-Brazaca Solange. Starch Digestibility and Physicochemical and Cooking Properties of Irradiated Rice Grains [J]. Rice Science, 2017, 24(1): 48-55. |
| [13] | S. Abe S., Yamasaki Y., Wakatsuki T.. Assessing Silicon Availability in Soils of Rice-Growing Lowlands and Neighboring Uplands in Benin and Nigeria [J]. Rice Science, 2016, 23(4): 196-202. |
| [14] | Saleethong Paweena, Roytrakul Sittiruk, Kong-Ngern Kanlaya, Theerakulpisut Piyada. Differential Proteins Expressed in Rice Leaves and Grains in Response to Salinity and Exogenous Spermidine Treatments [J]. Rice Science, 2016, 23(1): 9-21. |
| [15] | Hong-wei Zhang, Yu-yu Chen, Jun-yu Chen, Yu-jun Zhu, De-run Huang, Ye-yang Fan, Jie-yun Zhuang. Mapping of qTGW1.1, a Quantitative Trait Locus for 1000-Grain Weight in Rice (Oryza sativa L.) [J]. Rice Science, 2015, 22(1): 9-15. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||