
Rice Science ›› 2018, Vol. 25 ›› Issue (4): 175-184.DOI: 10.1016/j.rsci.2018.06.001
• Orginal Article • Next Articles
Fantao Zhang1( ), Yuan Luo1, Meng Zhang1, Yi Zhou1, Hongping Chen2, Biaolin Hu2, Jiankun Xie1(
), Yuan Luo1, Meng Zhang1, Yi Zhou1, Hongping Chen2, Biaolin Hu2, Jiankun Xie1( )
)
Received:2017-12-18
Accepted:2018-02-27
Online:2018-06-20
Published:2018-04-10
Fantao Zhang, Yuan Luo, Meng Zhang, Yi Zhou, Hongping Chen, Biaolin Hu, Jiankun Xie. Identification and Characterization of Drought Stress- Responsive Novel microRNAs in Dongxiang Wild Rice[J]. Rice Science, 2018, 25(4): 175-184.
Add to citation manager EndNote|Ris|BibTeX
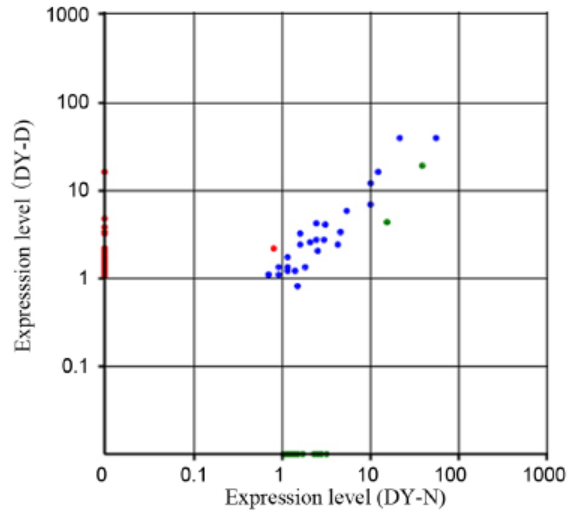
Fig. 1. Analysis of the differential expression of novel miRNAs. Each point represents one novel miRNA. Red points represent up-expressed miRNA with ratio > 2; blue points represent equally- expressed miRNA with 1/2 < ratio ≤ 2; green points represent down- expressed miRNA with ratio ≤ 1/2. Ratio = Normalized expression in the drought stress-treatment samples / Normalized expression in the control samples.
| Name | DY-N-std | DY-D-std | log2(DY-D-std/DY-N-std) | P-value | Up/down | Name | DY-N-std | DY-D-std | log2(DY-D-std/DY-N-std) | P-value | Up/down |
|---|---|---|---|---|---|---|---|---|---|---|---|
| novel-mir-135 | 3.26 | 0.01 | -8.35 | 2.77 × 10-8 | Down | novel-mir-68 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-96 | 2.79 | 0.01 | -8.13 | 3.36 × 10-7 | Down | novel-mir-79 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-115 | 2.56 | 0.01 | -8.00 | 1.17 × 10-6 | Down | novel-mir-82 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-125 | 2.33 | 0.01 | -7.86 | 4.08 × 10-6 | Down | novel-mir-85 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-134 | 1.75 | 0.01 | -7.45 | 9.23 × 10-5 | Down | novel-mir-12 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-94 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-24 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-103 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-29 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-111 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-36 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-98 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-38 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-114 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-57 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-137 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-66 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-88 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-73 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-99 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-4 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-116 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-44 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-89 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-46 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-105 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-48 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-110 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-70 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-122 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-40 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-129 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-41 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-133 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-60 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-93 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-17 | 0.01 | 1.61 | 7.33 | 9.29 × 10-5 | Up |
| novel-mir-97 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-74 | 0.01 | 1.61 | 7.33 | 9.29 × 10-5 | Up |
| novel-mir-100 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-33 | 0.01 | 1.75 | 7.45 | 4.31 × 10-5 | Up |
| novel-mir-108 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-55 | 0.01 | 1.75 | 7.45 | 4.31 × 10-5 | Up |
| novel-mir-130 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-34 | 0.01 | 1.88 | 7.55 | 2.00 × 10-5 | Up |
| novel-mir-3 | 15.59 | 4.30 | -1.86 | 3.27 × 10-13 | Down | novel-mir-26 | 0.01 | 2.01 | 7.65 | 9.29 × 10-6 | Up |
| novel-mir-69 | 38.63 | 18.80 | -1.04 | 1.16 × 10-13 | Down | novel-mir-45 | 0.01 | 2.15 | 7.75 | 4.31 × 10-6 | Up |
| novel-mir-5 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-54 | 0.01 | 2.15 | 7.75 | 4.31 × 10-6 | Up |
| novel-mir-7 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-84 | 0.01 | 3.22 | 8.33 | 9.30 × 10-9 | Up |
| novel-mir-10 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-2 | 0.01 | 3.36 | 8.39 | 4.32 × 10-9 | Up |
| novel-mir-42 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-51 | 0.01 | 3.76 | 8.55 | 4.32 × 10-10 | Up |
| novel-mir-50 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-6 | 0.01 | 4.70 | 8.88 | 2.00 × 10-12 | Up |
| novel-mir-52 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-71 | 0.01 | 16.12 | 10.65 | 9.35 × 10-41 | Up |
| novel-mir-61 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
Table 1 Drought stress-responsive novel miRNAs detected from Dongxiang wild rice (DXWR).
| Name | DY-N-std | DY-D-std | log2(DY-D-std/DY-N-std) | P-value | Up/down | Name | DY-N-std | DY-D-std | log2(DY-D-std/DY-N-std) | P-value | Up/down |
|---|---|---|---|---|---|---|---|---|---|---|---|
| novel-mir-135 | 3.26 | 0.01 | -8.35 | 2.77 × 10-8 | Down | novel-mir-68 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-96 | 2.79 | 0.01 | -8.13 | 3.36 × 10-7 | Down | novel-mir-79 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-115 | 2.56 | 0.01 | -8.00 | 1.17 × 10-6 | Down | novel-mir-82 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-125 | 2.33 | 0.01 | -7.86 | 4.08 × 10-6 | Down | novel-mir-85 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
| novel-mir-134 | 1.75 | 0.01 | -7.45 | 9.23 × 10-5 | Down | novel-mir-12 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-94 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-24 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-103 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-29 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-111 | 1.51 | 0.01 | -7.24 | 3.22 × 10-4 | Down | novel-mir-36 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-98 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-38 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-114 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-57 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-137 | 1.40 | 0.01 | -7.13 | 6.00 × 10-4 | Down | novel-mir-66 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-88 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-73 | 0.01 | 1.21 | 6.92 | 9.29 × 10-4 | Up |
| novel-mir-99 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-4 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-116 | 1.28 | 0.01 | -7.00 | 1.12 × 10-3 | Down | novel-mir-44 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-89 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-46 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-105 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-48 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-110 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-70 | 0.01 | 1.34 | 7.07 | 4.31 × 10-4 | Up |
| novel-mir-122 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-40 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-129 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-41 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-133 | 1.16 | 0.01 | -6.86 | 2.09 × 10-3 | Down | novel-mir-60 | 0.01 | 1.48 | 7.21 | 2.00 × 10-4 | Up |
| novel-mir-93 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-17 | 0.01 | 1.61 | 7.33 | 9.29 × 10-5 | Up |
| novel-mir-97 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-74 | 0.01 | 1.61 | 7.33 | 9.29 × 10-5 | Up |
| novel-mir-100 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-33 | 0.01 | 1.75 | 7.45 | 4.31 × 10-5 | Up |
| novel-mir-108 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-55 | 0.01 | 1.75 | 7.45 | 4.31 × 10-5 | Up |
| novel-mir-130 | 1.05 | 0.01 | -6.71 | 3.90 × 10-3 | Down | novel-mir-34 | 0.01 | 1.88 | 7.55 | 2.00 × 10-5 | Up |
| novel-mir-3 | 15.59 | 4.30 | -1.86 | 3.27 × 10-13 | Down | novel-mir-26 | 0.01 | 2.01 | 7.65 | 9.29 × 10-6 | Up |
| novel-mir-69 | 38.63 | 18.80 | -1.04 | 1.16 × 10-13 | Down | novel-mir-45 | 0.01 | 2.15 | 7.75 | 4.31 × 10-6 | Up |
| novel-mir-5 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-54 | 0.01 | 2.15 | 7.75 | 4.31 × 10-6 | Up |
| novel-mir-7 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-84 | 0.01 | 3.22 | 8.33 | 9.30 × 10-9 | Up |
| novel-mir-10 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-2 | 0.01 | 3.36 | 8.39 | 4.32 × 10-9 | Up |
| novel-mir-42 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-51 | 0.01 | 3.76 | 8.55 | 4.32 × 10-10 | Up |
| novel-mir-50 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-6 | 0.01 | 4.70 | 8.88 | 2.00 × 10-12 | Up |
| novel-mir-52 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up | novel-mir-71 | 0.01 | 16.12 | 10.65 | 9.35 × 10-41 | Up |
| novel-mir-61 | 0.01 | 1.07 | 6.75 | 2.00 × 10-3 | Up |
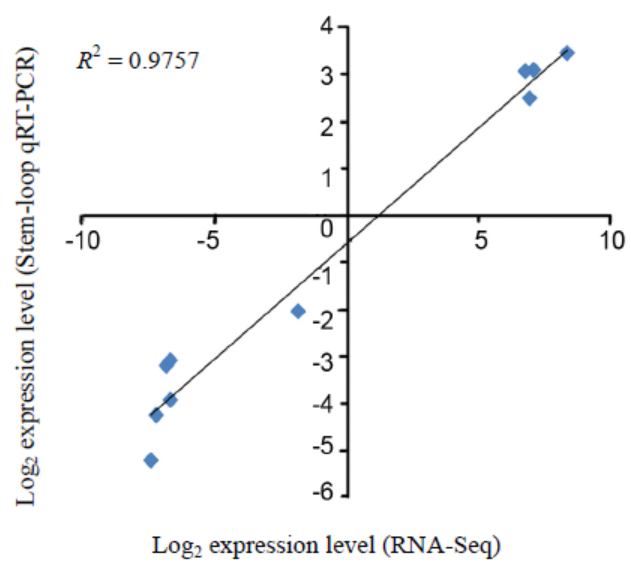
Fig. 2. Comparison of the expression levels of 10 randomly selected drought stress-responsive novel miRNAs using RNA-seq and stem-loop quantitative real-time PCR (qRT-PCR).Six down-regulated (novel-mir-3, novel-mir-93, novel-mir-97, novel-mir-103, novel-mir-105 and novel-mir-134) and four up- regulated (novel-mir-12, novel-mir-46, novel-mir-68 and novel-mir-84) miRNAs were used.
| [1] | Aravind J, Rinku S, Pooja B, Shikha M, Kaliyugam S, Mallikarjuna M G, Kumar A, Rao A R, Nepolean T.2017. Identification, characterization, and functional validation of drought-responsive microRNAs in subtropical maize inbreds.Front Plant Sci, 8: 941. |
| [2] | Arshad M, Feyissa B A, Amyot L, Aung B, Hannoufa A.2017. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci, 258: 122-136. |
| [3] | Baek D, Chun H J, Kang S, Shin G, Park S J, Hong H, Kim C, Kim D H, Lee S Y, Kim M C, Yun D J.2016. A role forArabidopsis miR399f in salt, drought, and ABA signaling. Mol Cells, 39(2): 111-118. |
| [4] | Cavrak V V, Lettner N, Jamge S, Kosarewicz A, Bayer L M, Scheid O M.2014. How a retrotransposon exploits the plant’s heat stress response for its activation.PLoS Genet, 10(1): e1004115. |
| [5] | Chen L H, Han J P, Deng X M, Tan S L, Li L L, Li L, Zhou J F, Peng H, Yang G X, He G Y, Zhang W X.2016. Expansion and stress responses of AP2/EREBP superfamily inBrachypodium distachyon. Sci Rep, 6: 21623. |
| [6] | Ding G H, Sun J, Yang G, Zhang F M, Bai L M, Sun S C, Jiang S K, Wang T T, Zheng H L, Xia T S, Shen X H, Ma D R, Chen W F.2016. Global genome expression analysis of root genes under drought stress in weedy rice and upland rice.Chin J Rice Sci, 30(5): 458-468. (in Chinese with English abstarct) |
| [7] | Fang Y J, Xie K B, Xiong L Z.2014. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice.J Exp Bot, 65(8): 2119-2135. |
| [8] | Fang Y J, Xiong L Z.2015. General mechanisms of drought response and their application in drought resistance improvement in plants.Cell Mol Life Sci, 72(4): 673-689. |
| [9] | Fukao T, Xiong L Z.2013. Genetic mechanisms conferring adaptation to submergence and drought in rice: Simple or complex?Curr Opin Plant Biol, 16(2): 196-204. |
| [10] | Guo Y Q, Zhao S S, Zhu C, Chang X J, Yue C, Wang Z, Lin Y L, Lai Z X.2017. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol, 17(1): 211. |
| [11] | Jain M, Nijhawan A, Tyagi A K, Khurana J P.2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR.Biochem Biophys Res Commun, 345(2): 646-651. |
| [12] | Jia Q S, Zhu J, Xu X F, Lou Y, Zhang Z L, Zhang Z P, Yang Z N.2015. Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for nexine formation.Mol Plant, 8(2): 251-260. |
| [13] | Jin Y, Luo Q, Tong H N, Wang A J, Cheng Z J, Tang J F, Li D Y, Zhao X F, Li X B, Wan J M, Jiao Y L, Chu C C, Zhu L H.2011. An AT-hook gene is required for palea formation and floral organ number control in rice.Dev Biol, 359(2): 277-288. |
| [14] | Kang J G, Guo Y Y, Chen Y J, Li H L, Zhang L, Liu H X.2014. Upregulation of the AT-hook DNA binding geneBoMF2 in OguCMS anthers of Brassica oleracea suggests that it encodes a transcriptional regulatory factor for anther development. Mol Biol Rep, 41(4): 2005-2014. |
| [15] | Khraiwesh B, Zhu J K, Zhu J H.2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants.Biochim Biophys Acta, 1819(2): 137-148. |
| [16] | Kim S Y, Kim Y C, Seong E S, Lee Y H, Park J M, Choi D.2007. The chili pepper CaATL1: An AT-hook motif-containing transcription factor implicated in defence responses against pathogens.Mol Plant Pathol, 8(6): 761-771. |
| [17] | Krannich C T, Maletzki L, Kurowsky C, Horn R.2015. Network candidate genes in breeding for drought tolerant crops.Int J Mol Sci, 16(7): 16378-16400. |
| [18] | Lesk C, Rowhani P, Ramankutty N.2016. Influence of extreme weather disasters on global crop production. Nature, 529: 84-87. |
| [19] | Li H, Wang Y, Wu M, Li L H, Jin C, Zhang Q L, Chen C B, Song W Q, Wang C G.2017. Small RNA sequencing reveals differential miRNA expression in the early development of Broccoli (Brassica oleracea var. italica) pollen. Front Plant Sci, 8: 404. |
| [20] | Li J F, Yang J C.2017. Research advances in the effects of water, nitrogen and their interaction on the yield, water and nitrogen use efficiencies of rice.Chin J Rice Sci, 31(3): 327-334. (in Chinese with English abstarct) |
| [21] | Li W B, Wang T, Zhang Y H, Li Y G.2016. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenicArabidopsis thaliana. J Exp Bot, 67(1): 175-194. |
| [22] | Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B R.2012. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucl Acids Res, 40(10): 4298-4305. |
| [23] | Lian S, Cho W K, Kim S M, Choi H, Kim K H.2016. Time-course small RNA profiling reveals rice miRNAs and their target genes in response to rice stripe virus infection.PLoS One, 11(9): e0162319. |
| [24] | Lim L P, Glasner M E, Yekta S, Burge C B, Bartel D P.2003. Vertebrate microRNA genes.Science, 299: 1540. |
| [25] | Liu M M, Yu H Y, Zhao G J, Huang Q F, Lu Y G, Ouyang B.2018. Identification of drought-responsive microRNAs in tomato using high-throughput sequencing.Funct Integr Genom, 18(1): 67-78. |
| [26] | Liu Q L, Xu K D, Zhong M, Pan Y Z, Jiang B B, Liu G L, Jia Y, Zhang H Q.2013. Overexpression of a novel chrysanthemum Cys2/His2-type zinc finger protein gene DgZFP3 confers drought tolerance in tobacco. Biotechnol Lett, 35(11): 1953-1959. |
| [27] | Lu X K, Chen X G, Mu M, Wang J J, Wang X G, Wang D L, Yin Z J, Fan W L, Wang S, Guo L X, Ye W W.2016. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS One, 11(6): e0156723. |
| [28] | Luo L J.2010. Breeding for water-saving and drought-resistance rice (WDR) in China.J Exp Bot, 61(13): 3509-3517. |
| [29] | Luo X, Bai X, Zhu D, Li Y, Ji W, Cai H, Wu J, Liu B H, Zhu Y M.2012. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress.Planta, 235(6): 1141-1155. |
| [30] | Mao D H, Yu L, Chen D Z, Li L Y, Zhu Y X, Xiao Y Q, Zhang D C, Chen C Y.2015. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet, 128(7): 1359-1371. |
| [31] | Pandey V, Shukla A.2015. Acclimation and tolerance strategies of rice under drought stress.Rice Sci, 22(4): 147-161. |
| [32] | Pei H X, Ma N, Chen J W, Zheng Y, Tian J, Li J, Zhang S, Fei Z J, Gao J P.2013. Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening.PLoS One, 8(5): e64290. |
| [33] | Sailaja B, Voleti S R, Subrahmanyam D, Sarla N, Prasanth V V, Bhadana V P, Mangrauthia S K.2014. Prediction and expression analysis of miRNAs associated with heat stress inOryza sativa. Rice Sci, 21(1): 3-12. |
| [34] | Shriram V, Kumar V, Devarumath R M, Khare T S, Wani S H.2016. MicroRNAs as potential targets for abiotic stress tolerance in plants.Front Plant Sci, 7: 817. |
| [35] | Song J B, Gao S, Sun D, Li H, Shu X X, Yang Z M.2013. miR394 and LCR are involved inArabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol, 13: 210. |
| [36] | Song X W, Cao X F.2017. Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptation in rice.Curr Opin Plant Biol, 36: 111-118. |
| [37] | Tang J Y, Chu C C.2017. MicroRNAs in crop improvement: Fine-tuners for complex traits.Nat Plants, 3(7): 17077. |
| [38] | Tian X H, Li X P, Zhou H L, Zhang J S, Gong Z Z, Chen S Y.2005. OsDREB4 genes in rice encode AP2-containing proteins that bind specifically to the dehydration-responsive element.J Integr Plant Biol, 47(4): 467-476. |
| [39] | Torres-Franklin M L, Repellin A, Huynh V B, D’Arcy-Lameta A, Zuily-Fodil Y, Pham-Thi A T.2009. Omega-3 fatty acid desaturase (FAD3, FAD7, FAD8) gene expression and linolenic acid content in cowpea leaves submitted to drought and after rehydration.Environ Exp Bot, 65: 162-169. |
| [40] | Wang C, Han J, Liu C H, Kibet K N, Kayesh E, Shangguan L F, Li X Y, Fang J G.2012. Identification of microRNAs from amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genom, 13: 122. |
| [41] | Xia K F, Wang R, Ou X J, Fang Z M, Tian C, Duan J, Wang Y Q, Zhang M Y.2012. OsTIR1 andOsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One, 7(1): e30039. |
| [42] | Xing L B, Zhang D, Zhao C P, Li Y M, Ma J J, An N, Han M Y.2016. Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domestica Borkh.). Plant Biotechnol J, 14(2): 749-770. |
| [43] | Yan J, Zhao C Z, Zhou J P, Yang Y, Wang P C, Zhu X H, Tang G L, Bressan R A, Zhu J K.2016. The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response inArabidopsis thaliana. PLoS Genet, 12(11): e1006416. |
| [44] | Yasuda K, Ito M, Sugita T, Tsukiyama T, Saito H, Naito K, Teraishi M, Tanisaka T, Okumoto Y.2013. Utilization of transposable element as a novel genetic tool for modification of the stress response in rice. Mol Breeding, 32(3): 505-516. |
| [45] | Zhang F T, Luo X D, Hu B L, Wan Y, Xie J K.2013. YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice.Rice, 6(1): 7. |
| [46] | Zhang F T, Luo X D, Zhou Y, Xie J K.2016. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol Lett, 38(4): 711-721. |
| [47] | Zhang H, Liu Y P, Wen F, Yao D M, Wang L, Guo J, Ni L, Zhang A Y, Tan M P, Jiang M Y.2014. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice.J Exp Bot, 65(20): 5795-5809. |
| [48] | Zhang J W, Long Y, Xue M D, Xiao X G, Pei X W.2017. Identification of microRNAs in response to drought in common wild rice (Oryza rufipogon Griff.) shoots and roots. PLoS One, 12(1): e0170330. |
| [49] | Zhang J Y, Broeckling C D, Blancaflor E B, Sledge M K, Sumner L W, Wang Z Y.2005. Overexpression ofWXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa(Medicago sativa). Plant J, 42(5): 689-707. |
| [50] | Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin-Frenkel A, Salts Y, Shabtai S, Ben-Hayyim G.2005. Modulated fatty acid desaturation via overexpression of two distinct ω-3 desaturases differentially alters tolerance tovarious abiotic stresses in transgenic tobacco cells and plants.Plant J, 44(3): 361-371. |
| [51] | Zhang X, Zhou S X, Fu Y C, Su Z, Wang X K, Sun C Q.2006. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol Biol, 62(1/2): 247-259. |
| [52] | Zhao L F, Hu Y B, Chong K, Wang T.2010. ARAG1, an ABA- responsive DREB gene, plays a role in seed germination and drought tolerance of rice.Ann Bot, 105(3): 401-409. |
| [53] | Zhou L G, Liu Y H, Liu Z C, Kong D Y, Duan M, Luo L J.2010. Genome-wide identification and analysis of drought-responsive microRNAs inOryza sativa. J Exp Bot, 61(15): 4157-4168. |
| [54] | Zhou L G, Liu Z C, Liu Y H, Kong D Y, Li T F, Yu S W, Mei H W, Xu X Y, Liu H Y, Chen L, Luo L J.2016. A novel geneOsAHL1 improves both drought avoidance and drought tolerance in rice. Sci Rep, 6: 30264. |
| [1] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [2] | Chen Eryong, Shen Bo. OsABT, a Rice WD40 Domain-Containing Protein, Is Involved in Abiotic Stress Tolerance [J]. Rice Science, 2022, 29(3): 247-256. |
| [3] | Nie Yuanyuan, Xia Hui, Ma Xiaosong, Lou Qiaojun, Liu Yi, Zhang Anling, Cheng Liang, Yan Longan, Luo Lijun. Dissecting Genetic Basis of Deep Rooting in Dongxiang Wild Rice [J]. Rice Science, 2022, 29(3): 277-287. |
| [4] | Muhammad Mahran Aslam, Muhammad Abdul Rehman Rashid, Mohammad Aquil Siddiqui, Muhammad Tahir Khan, Fozia Farhat, Shafquat Yasmeen, Imtiaz Ahmad Khan, Shameem Raja, Fatima Rasool, Mahboob Ali Sial, Zhao Yan. Recent Insights into Signaling Responses to Cope Drought Stress in Rice [J]. Rice Science, 2022, 29(2): 105-117. |
| [5] | Balija Vishalakshi, Bangale Umakanth, Ponnuvel Senguttuvel, Makarand Barbadikar Kalyani, Prasad Madamshetty Srinivas, Rao Durbha Sanjeeva, Yadla Hari, Madhav Maganti Sheshu. Improvement of Upland Rice Variety by Pyramiding Drought Tolerance QTL with Two Major Blast Resistance Genes for Sustainable Rice Production [J]. Rice Science, 2021, 28(5): 493-500. |
| [6] | Vera Jesus Da Costa Maria, Ramegowda Yamunarani, Ramegowda Venkategowda, N. Karaba Nataraja, M. Sreeman Sheshshayee, Udayakumar Makarla. Combined Drought and Heat Stress in Rice: Responses, Phenotyping and Strategies to Improve Tolerance [J]. Rice Science, 2021, 28(3): 233-242. |
| [7] | Baoxiang Wang, Yan Liu, Yifeng Wang, Jingfang Li, Zhiguang Sun, Ming Chi, Yungao Xing, Bo Xu, Bo Yang, Jian Li, Jinbo Liu, Tingmu Chen, Zhaowei Fang, Baiguan Lu, Dayong Xu, Kazeem Bello Babatunde. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1 [J]. Rice Science, 2021, 28(3): 257-267. |
| [8] | Panda Debabrata, Sakambari Mishra Swati, Kumar Behera Prafulla. Drought Tolerance in Rice: Focus on Recent Mechanisms and Approaches [J]. Rice Science, 2021, 28(2): 119-132. |
| [9] | Weidong Qi, Hongping Chen, Zuozhen Yang, Biaolin Hu, Xiangdong Luo, Bing Ai, Yuan Luo, Yu Huang, Jiankun Xie, Fantao Zhang. Systematic Characterization of Long Non-Coding RNAs and Their Responses to Drought Stress in Dongxiang Wild Rice [J]. Rice Science, 2020, 27(1): 21-31. |
| [10] | Donde Ravindra, Kumar Jitendra, Gouda Gayatri, Kumar Gupta Manoj, Mukherjee Mitadru, Yasin Baksh Sk, Mahadani Pradosh, Kumar Sahoo Khirod, Behera Lambodar, Kumar Dash Sushanta. Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers [J]. Rice Science, 2019, 26(4): 239-247. |
| [11] | Nahar Shamsun, R. Vemireddy Lakshminarayana, Sahoo Lingaraj, Tanti Bhaben. Antioxidant Protection Mechanisms Reveal Significant Response in Drought-Induced Oxidative Stress in Some Traditional Rice of Assam, India [J]. Rice Science, 2018, 25(4): 185-196. |
| [12] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [13] | Sakambari Mishra Swati, Panda Debabrata. Leaf Traits and Antioxidant Defense for Drought Tolerance During Early Growth Stage in Some Popular Traditional Rice Landraces from Koraput, India [J]. Rice Science, 2017, 24(4): 207-217. |
| [14] | Pandey Veena, Shukla Alok. Acclimation and Tolerance Strategies of Rice under Drought Stress [J]. Rice Science, 2015, 22(4): 147-161. |
| [15] | ZHENG Xiao-guo, CHEN Liang, LOU Qiao-jun, XIA Hui, LI Ming-shou, LUO Li-jun. Changes in DNA Methylation Pattern at Two Seedling Stages in Water Saving and Drought-Resistant Rice Variety after Drought Stress Domestication [J]. RICE SCIENCE, 2014, 21(5): 262-270. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||