
Rice Science ›› 2020, Vol. 27 ›› Issue (1): 21-31.DOI: 10.1016/j.rsci.2019.12.003
• Research Paper • Previous Articles Next Articles
Weidong Qi1, Hongping Chen2, Zuozhen Yang3, Biaolin Hu2, Xiangdong Luo1, Bing Ai1, Yuan Luo1, Yu Huang1, Jiankun Xie1( ), Fantao Zhang1(
), Fantao Zhang1( )
)
Received:2018-07-20
Accepted:2018-09-27
Online:2020-01-28
Published:2019-09-30
About author: These authors contributed equally to this work
Weidong Qi, Hongping Chen, Zuozhen Yang, Biaolin Hu, Xiangdong Luo, Bing Ai, Yuan Luo, Yu Huang, Jiankun Xie, Fantao Zhang. Systematic Characterization of Long Non-Coding RNAs and Their Responses to Drought Stress in Dongxiang Wild Rice[J]. Rice Science, 2020, 27(1): 21-31.
Add to citation manager EndNote|Ris|BibTeX
| Parameter | Control (DY-CK) | Drought stress (DY-D) |
|---|---|---|
| Raw base number | 17 474 692 200 | 15 504 071 700 |
| Clean base number | 13 883 436 900 | 12 486 326 400 |
| Raw read number | 116 497 948 | 103 360 478 |
| Low-quality read number a | 9 723 402 (8.35%) | 8 005 290 (7.75%) |
| Adapter polluted read number a | 10 420 480 (8.95%) | 8 331 824 (8.06%) |
| Clean read number a | 92 556 246 (79.45%) | 83 242 176 (80.54%) |
| Unmapped reads b | 4 598 146 (4.97%) | 4 835 398 (5.81%) |
| Total mapped reads b | 87 958 100 (95.03%) | 78 406 778 (94.19%) |
| Multiple mapped reads b | 31 889 847 (34.45%) | 21 343 543 (25.64%) |
| Unique mapped reads b | 56 068 253 (60.58%) | 57 063 235 (68.55%) |
| Reads map to exonic region (%) | 66.01 | 71.79 |
| Reads map to intronic region (%) | 6.58 | 6.05 |
| Reads map to intergenic region (%) | 27.41 | 22.16 |
Table 1 RNA sequencing data for two samples.
| Parameter | Control (DY-CK) | Drought stress (DY-D) |
|---|---|---|
| Raw base number | 17 474 692 200 | 15 504 071 700 |
| Clean base number | 13 883 436 900 | 12 486 326 400 |
| Raw read number | 116 497 948 | 103 360 478 |
| Low-quality read number a | 9 723 402 (8.35%) | 8 005 290 (7.75%) |
| Adapter polluted read number a | 10 420 480 (8.95%) | 8 331 824 (8.06%) |
| Clean read number a | 92 556 246 (79.45%) | 83 242 176 (80.54%) |
| Unmapped reads b | 4 598 146 (4.97%) | 4 835 398 (5.81%) |
| Total mapped reads b | 87 958 100 (95.03%) | 78 406 778 (94.19%) |
| Multiple mapped reads b | 31 889 847 (34.45%) | 21 343 543 (25.64%) |
| Unique mapped reads b | 56 068 253 (60.58%) | 57 063 235 (68.55%) |
| Reads map to exonic region (%) | 66.01 | 71.79 |
| Reads map to intronic region (%) | 6.58 | 6.05 |
| Reads map to intergenic region (%) | 27.41 | 22.16 |
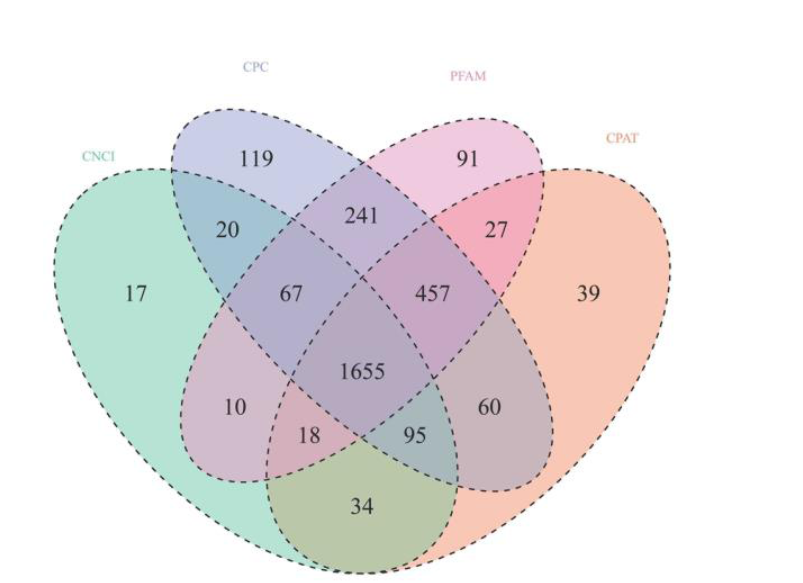
Fig. 1. Venn diagram of non-coding transcripts from the Coding-Non-Coding Index (CNCI), the Coding Potential Calculator (CPC), the Protein Families Database (PFAM), and the Coding-Potential Assessment Tool (CPAT).
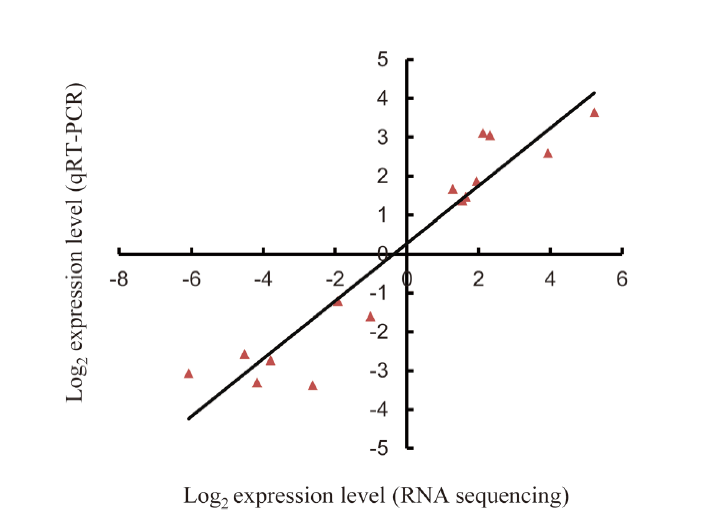
Fig. 2. Comparison of expression levels of 15 randomly selected drought responsive lncRNAs using RNA sequencing and qRT-PCR methods. ===Seven down-regulated (MSTRG25356, MSTRG29971, MSTRG42925, MSTRG45354, MSTRG54123, MSTRG55730 and MSTRG69646) and eight up-regulated (MSTRG5459, MSTRG14660, MSTRG21367, MSTRG22525, MSTRG31874, MSTRG43289, MSTRG46940 and MSTRG62647) lncRNAs were used.
| [1] | Ahmad I, Devonshire J, Mohamed R, Schultze M, Maathuis F J M.2016. Overexpression of the potassium channel TPKb in small vacuoles confers osmotic and drought tolerance to rice.New Phytol, 209(3): 1040-1048. |
| [2] | Anupama A, Bhugra S, Lall B, Chaudhury S, Chugh A.2018. Assessing the correlation of genotypic and phenotypic responses of indica rice varieties under drought stress. Plant Physiol Biochem, 127: 343-354. |
| [3] | Cagirici H B, Alptekin B, Budak H.2017. RNA sequencing and co-expressed long non-coding RNA in modern and wild wheats. Sci Rep, 7(1): 10670. |
| [4] | Cao J N.2014. The functional role of long non-coding RNAs and epigenetics.Biol Proced Online, 16: 11. |
| [5] | Dröge-Laser W, Snoek B L, Snel B, Weiste C.2018. TheArabidopsis bZIP transcription factor family: An update. Curr Opin Plant Biol, 45: 36-49. |
| [6] | Duan H, Tong H, Liu Y Q, Xu Q F, Ma J, Wang C M.2019. Research advances in the effect of heat and drought on rice and its mechanism.Chin J Rice Sci, 33(3): 206-218. (in Chinese with English abstract) |
| [7] | Heo J B, Lee Y S, Sung S.2013. Epigenetic regulation by long noncoding RNAs in plants.Chrom Res, 21: 685-693. |
| [8] | Huang L Y, Zhang F, Zhang F, Wang W S, Zhou Y L, Fu B Y, Li Z K.2014. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress.BMC Genom, 15(1): 1026. |
| [9] | Jain M, Nijhawan A, Tyagi A K, Khurana J P.2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR.Biochem Biophys Res Commun, 345(2): 646-651. |
| [10] | Jisha V, Dampanaboina L, Vadassery J, Mithöfer A, Kappara S, Ramanan R.2015. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS One, 10(6): e0127831. |
| [11] | Joshi R, Wani S H, Singh B, Bohra A, Dar Z A, Lone A A, Pareek A, Singla-Pareek S L.2016. Transcription factors and plants response to drought stress: Current understanding and future directions.Front Plant Sci, 7: 1029. |
| [12] | Kim D, Langmead B, Salzberg S L.2015. HISAT: A fast spliced aligner with low memory requirements.Nat Methods, 12(4): 357-360. |
| [13] | Kiranmai K, Lokanadha Rao G, Pandurangaiah M, Nareshkumar A, Amaranatha Reddy V, Lokesh U, Venkatesh B, Anthony Johnson A M, Sudhakar C.2018. A novel WRKY transcription factor,MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut(Arachis hypogaea L.) plants. Front Plant Sci, 9: 346. |
| [14] | Kong L, Zhang Y, Ye Z Q, Liu X Q, Zhao S Q, Wei L P, Gao G.2007. Cpc: Assess the protein-coding potential of transcripts using sequence features and support vector machine.Nucl Acids Res, 35: W345-W349. |
| [15] | Kroll K W, Mokaram N E, Pelletier A R, Frankhouser D E, Westphal M S, Stump P A, Stump C L, Bundschuh R, Blachly J S, Yan P.2014. Quality control for RNA-Seq (QuaCRS): An integrated quality control pipeline.Cancer Inform, 13: 7-14. |
| [16] | Lee D K, Kim H I, Jang G, Chung P J, Jeong J S, Kim Y S, Bang S W, Jung H, Choi Y D, Kim J K.2015. The NF-YA transcription factorOsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci, 241: 199-210. |
| [17] | Li L, Eichten S R, Shimizu R, Petsch K, Yeh C T, Wu W, Chettoor A M, Givan S A, Cole R A, Fowler J E, Evans M M S, Scanlon M J, Yu J M, Schnable P S, Timmermans M C P, Springer N M, Muehlbauer G J.2014. Genome-wide discovery and characterization of maize long non-coding RNAs.Genom Biol, 15: R40. |
| [18] | Li S J, Castillo-González C, Yu B, Zhang X R.2017. The functions of plant small RNAs in development and in stress responses.Plant J, 90: 654-670. |
| [19] | Li S X, Yu X, Lei N, Cheng Z H, Zhao P J, He Y K, Wang W Q, Peng M.2017. Corrigendum: Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep, 7: 46795. |
| [20] | Liang Y S, Zheng J, Yan C, Li X X, Liu S F, Zhou J J, Qin X J, Nan W B, Yang Y Q, Zhang H M.2018. Locating QTLs controlling overwintering trait in Chinese perennial Dongxiang wild rice.Mol Genet Genom, 293(1): 81-93. |
| [21] | Liu H P, Yu H Y, Tang G L, Huang T B.2018. Small but powerful: Function of microRNAs in plant development.Plant Cell Rep, 37(3): 515-528. |
| [22] | Lu X K, Chen X G, Mu M, Wang J J, Wang X G, Wang D L, Yin Z J, Fan W L, Wang S, Guo L X, Ye W W.2016. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS One, 11(6): e0156723. |
| [23] | Luo L J.2010. Breeding for water-saving and drought-resistance rice (WDR) in China.J Exp Bot, 61(13): 3509-3517. |
| [24] | Luo X, Bai X, Zhu D, Li Y, Ji W, Cai H, Wu J, Liu B H, Zhu Y M.2012. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress.Planta, 235(6): 1141-1155. |
| [25] | Ma J H, Gao X L, Liu Q, Shao Y, Zhang D J, Jiang L N, Li C X.2017. Overexpression ofTaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front Plant Sci, 8: 2036. |
| [26] | Mao X Z, Cai T, Olyarchuk J G, Wei L P.2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary.Bioinformatics, 21(19): 3787-3793. |
| [27] | Menguer P K, Sperotto R A, Ricachenevsky F K.2017. A walk on the wild side:Oryza species as source for rice abiotic stress tolerance. Genet Mol Biol, 40: 238-252. |
| [28] | Moran E, Lauder J, Musser C, Stathos A, Shu M J.2017. The genetics of drought tolerance in conifers.New Phytol, 216(4): 1034-1048. |
| [29] | Munné-Bosch S, Falara V, Pateraki I, López-Carbonell M, Cela J, Kanellis A K.2009. Physiological and molecular responses of the isoprenoid biosynthetic pathway in a drought-resistant Mediterranean shrub,Cistus creticus exposed to water deficit. J Plant Physiol, 166(2): 136-145. |
| [30] | Nawaz F, Naeem M, Zulfiqar B, Akram A, Ashraf M Y, Raheel M, Shabbir R N, Hussain R A, Anwar I, Aurangzaib M.2017. Understanding brassinosteroid-regulated mechanisms to improve stress tolerance in plants: A critical review.Environ Sci Pollut Res Int, 24(19): 15959-15975. |
| [31] | Pauli A, Valen E, Lin M F, Garber M, Vastenhouw N L, Levin J Z, Fan L, Sandelin A, Rinn J L, Regev A, Schier A F.2012. Systematic identifcation of long noncoding RNAs expressed during zebrafsh embryogenesis.Genome Res, 22(3): 577-591. |
| [32] | Pertea M, Pertea G M, Antonescu C M, Chang T C, Mendell J T, Salzberg S L.2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads.Nat Biotechnol, 33(3): 290-295. |
| [33] | Phukan U J, Jeena G S, Shukla R K.2016. WRKY transcription factors: Molecular regulation and stress responses in plants.Front Plant Sci, 7: 760. |
| [34] | Punta M, Coggill P C, Eberhardt R Y, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer E L L, Eddy S R, Bateman A, Finn R D.2012. The Pfam protein families database.Nucl Acids Res, 40: D290-D301. |
| [35] | Qin T, Zhao H Y, Cui P, Albesher N, Xiong L M.2017. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance.Plant Physiol, 175(3): 1321-1336. |
| [36] | Seo J S, Joo J, Kim M J, Kim Y K, Nahm B H, Song S I, Cheong J J, Lee J S, Kim J K, Choi Y D.2011. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice.Plant J, 65(6): 907-921. |
| [37] | Shanker A K, Maheswari M, Yadav S K, Desai S, Bhanu D, Attal N B, Venkateswarlu B.2014. Drought stress responses in crops.Funct Integr Genom, 14(1): 11-22. |
| [38] | Song X W, Cao X F.2017. Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptation in rice.Curr Opin Plant Biol, 36: 111-118. |
| [39] | Song Y, Jing S J, Yu D Q.2009. Overexpression of the stress- inducedOsWRKY08 improves osmotic stress tolerance in Arabidopsis. Chin Sci Bull, 54(24): 4671-4678. |
| [40] | Song Y, Chen L G, Zhang L P, Yu D Q.2010. Overexpression ofOsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis.J Biosci, 35(3): 459-471. |
| [41] | Sun L, Luo H T, Bu D C, Zhao G G, Yu K T, Zhang C H, Liu Y N, Chen R S, Zhao Y.2013. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts.Nucl Acids Res, 41: e166. |
| [42] | Sun X, Zhang Y P, Zhu X D, Korir N K, Tao R, Wang C, Fang J G.2014. Advances in identification and validation of plant microRNAs and their target genes.Physiol Plant, 152(2): 203-218. |
| [43] | Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, van Baren M J, Salzberg S L, Wold B J, Pachter L.2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation.Nat Biotechnol, 28: 511-515. |
| [44] | Wang L G, Park H J, Dasari S, Wang S Q, Kocher J P, Li W.2013. Cpat: Coding-potential assessment tool using an alignment-free logistic regression model.Nucl Acids Res, 41(6): e74. |
| [45] | Wang L K, Feng Z X, Wang X, Wang X W, Zhang X G.2010. Degseq: An R package for identifying differentially expressed genes from RNA-seq data.Bioinformatics, 26(1): 136-138. |
| [46] | Wang L L, Yu C C, Chen C, He C L, Zhu Y G, Huang W C.2014. Identification of rice Di19 family reveals OsDi19-4 involved in drought resistance. Plant Cell Rep, 33(12): 2047-2062. |
| [47] | Xiong L Z, Yang Y N.2003. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid- inducible mitogen-activated protein kinase.Plant Cell, 15(3): 745-759. |
| [48] | Xu D Q, Huang J, Guo S Q, Yang X, Bao Y M, Tang H J, Zhang H S.2008. Overexpression of a TFIIIA-type zinc finger protein geneZFP252 enhances drought and salt tolerance in rice(Oryza sativa L.). FEBS Lett, 582(7): 1037-1043. |
| [49] | Yang J, Chen X R, Zhu C L, Peng X S, He X P, Fu J R, Ouyang L J, Bian J M, Hu L F, Sun X T, Xu J, He H H.2015. RNA-seq reveals differentially expressed genes of rice (Oryza sativa) spikelet in response to temperature interacting with nitrogen at meiosis stage. BMC Genom, 16: 959. |
| [50] | Yasuda K, Ito M, Sugita T, Tsukiyama T, Saito H, Naito K, Teraishi M, Tanisaka T, Okumoto Y.2013. Utilization of transposable element as a novel genetic tool for modification of the stress response in rice.Mol Breeding, 32(3): 505-516. |
| [51] | Ye N H, Zhu G H, Liu Y G, Li Y X, Zhang J H.2011. ABA controls H2O2 accumulation through the induction ofOsCATB in rice leaves under water stress. Plant Cell Physiol, 52(4): 689-698. |
| [52] | Yin M Z, Wang Y P, Zhang L H, Li J Z, Quan W L, Yang L, Wang Q F, Chan Z L.2017. TheArabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J Exp Bot, 68(11): 2991-3005. |
| [53] | Young M D, Wakefield M J, Smyth G K, Oshlack A.2010. Gene ontology analysis for RNA-seq: Accounting for selection bias.Genom Biol, 11(2): R14. |
| [54] | Yu X, Niu X L, Yang S H, Li Y X, Liu L L, Tang W, Liu Y S.2011. Research on heat and drought tolerance in rice (Oryza sativa L.) by overexpressing transcription factor OsbZIP60. Sci Agric Sin, 44(20): 4142-4149. |
| [55] | Yuan J P, Li J R, Yang Y, Tan C, Zhu Y M, Hu L, Qi Y J, Lu Z J.2018. Stress-responsive regulation of long non-coding RNA polyadenylation inOryza sativa. Plant J, 93(5): 814-827. |
| [56] | Zaynab M, Fatima M, Abbas S, Umair M, Sharif Y, Raza M A.2018. Long non-coding RNAs as molecular players in plant defense against pathogens.Microb Pathog, 121: 277-282. |
| [57] | Zhang F T, Luo X D, Zhou Y, Xie J K.2016. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol Lett, 38(4): 711-721. |
| [58] | Zhang F T, Luo Y, Zhang M, Zhou Y, Chen H P, Hu B L, Xie J K.2018. Identification and characterization of drought stress- responsive novel microRNAs in Dongxiang wild rice.Rice Sci, 25(4): 175-184. |
| [59] | Zhang G Y, Duan A G, Zhang J G, He C Y.2017. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit. Gene, 596: 130-136. |
| [60] | Zhang L Y, Gu L K, Ringler P, Smith S, Rushton P J, Shen Q J.2015. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells.Plant Sci, 236: 214-222. |
| [61] | Zhang W, Han Z X, Guo Q L, Liu Y, Zheng Y X, Wu F L, Jin W B.2014. Identification of maize long non-coding RNAs responsive to drought stress.PLoS One, 9(6): e98958. |
| [62] | Zhang X, Zhou S X, Fu Y C, Su Z, Wang X K, Sun C Q.2006. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol Biol, 62: 247-259. |
| [63] | Zhao J F, Favero D S, Qiu J W, Roalson E H, Neff M M.2014. Insights into the evolution and diversification of the AT-hook motif nuclear localized gene family in land plants. BMC Plant Biol, 14: 266. |
| [64] | Zhou L G, Liu Z C, Liu Y H, Kong D Y, Li T F, Yu S W, Mei H W, Xu X Y, Liu H Y, Chen L, Luo L J.2016. A novel geneOsAHL1 improves both drought avoidance and drought tolerance in rice. Sci Rep, 6: 30264. |
| [65] | Zhu B Z, Yang Y F, Li R, Fu D Q, Wen L W, Luo Y B, Zhu H L.2015. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening.J Exp Bot, 66(15): 4483-4495. |
| [1] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [2] | Chen Eryong, Shen Bo. OsABT, a Rice WD40 Domain-Containing Protein, Is Involved in Abiotic Stress Tolerance [J]. Rice Science, 2022, 29(3): 247-256. |
| [3] | Nie Yuanyuan, Xia Hui, Ma Xiaosong, Lou Qiaojun, Liu Yi, Zhang Anling, Cheng Liang, Yan Longan, Luo Lijun. Dissecting Genetic Basis of Deep Rooting in Dongxiang Wild Rice [J]. Rice Science, 2022, 29(3): 277-287. |
| [4] | Muhammad Mahran Aslam, Muhammad Abdul Rehman Rashid, Mohammad Aquil Siddiqui, Muhammad Tahir Khan, Fozia Farhat, Shafquat Yasmeen, Imtiaz Ahmad Khan, Shameem Raja, Fatima Rasool, Mahboob Ali Sial, Zhao Yan. Recent Insights into Signaling Responses to Cope Drought Stress in Rice [J]. Rice Science, 2022, 29(2): 105-117. |
| [5] | Balija Vishalakshi, Bangale Umakanth, Ponnuvel Senguttuvel, Makarand Barbadikar Kalyani, Prasad Madamshetty Srinivas, Rao Durbha Sanjeeva, Yadla Hari, Madhav Maganti Sheshu. Improvement of Upland Rice Variety by Pyramiding Drought Tolerance QTL with Two Major Blast Resistance Genes for Sustainable Rice Production [J]. Rice Science, 2021, 28(5): 493-500. |
| [6] | Vera Jesus Da Costa Maria, Ramegowda Yamunarani, Ramegowda Venkategowda, N. Karaba Nataraja, M. Sreeman Sheshshayee, Udayakumar Makarla. Combined Drought and Heat Stress in Rice: Responses, Phenotyping and Strategies to Improve Tolerance [J]. Rice Science, 2021, 28(3): 233-242. |
| [7] | Baoxiang Wang, Yan Liu, Yifeng Wang, Jingfang Li, Zhiguang Sun, Ming Chi, Yungao Xing, Bo Xu, Bo Yang, Jian Li, Jinbo Liu, Tingmu Chen, Zhaowei Fang, Baiguan Lu, Dayong Xu, Kazeem Bello Babatunde. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1 [J]. Rice Science, 2021, 28(3): 257-267. |
| [8] | Panda Debabrata, Sakambari Mishra Swati, Kumar Behera Prafulla. Drought Tolerance in Rice: Focus on Recent Mechanisms and Approaches [J]. Rice Science, 2021, 28(2): 119-132. |
| [9] | Caixia Gao, Xiuwen Zheng, Hubo Li, Ali Mlekwa Ussi, Yu Gao, Jie Xiong. Roles of lncRNAs in Rice: Advances and Challenges [J]. Rice Science, 2020, 27(5): 384-395. |
| [10] | Donde Ravindra, Kumar Jitendra, Gouda Gayatri, Kumar Gupta Manoj, Mukherjee Mitadru, Yasin Baksh Sk, Mahadani Pradosh, Kumar Sahoo Khirod, Behera Lambodar, Kumar Dash Sushanta. Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers [J]. Rice Science, 2019, 26(4): 239-247. |
| [11] | Fantao Zhang, Yuan Luo, Meng Zhang, Yi Zhou, Hongping Chen, Biaolin Hu, Jiankun Xie. Identification and Characterization of Drought Stress- Responsive Novel microRNAs in Dongxiang Wild Rice [J]. Rice Science, 2018, 25(4): 175-184. |
| [12] | Nahar Shamsun, R. Vemireddy Lakshminarayana, Sahoo Lingaraj, Tanti Bhaben. Antioxidant Protection Mechanisms Reveal Significant Response in Drought-Induced Oxidative Stress in Some Traditional Rice of Assam, India [J]. Rice Science, 2018, 25(4): 185-196. |
| [13] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [14] | Sakambari Mishra Swati, Panda Debabrata. Leaf Traits and Antioxidant Defense for Drought Tolerance During Early Growth Stage in Some Popular Traditional Rice Landraces from Koraput, India [J]. Rice Science, 2017, 24(4): 207-217. |
| [15] | Pandey Veena, Shukla Alok. Acclimation and Tolerance Strategies of Rice under Drought Stress [J]. Rice Science, 2015, 22(4): 147-161. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||