
Rice Science ›› 2022, Vol. 29 ›› Issue (3): 237-246.DOI: 10.1016/j.rsci.2022.01.010
• Research Paper • Previous Articles Next Articles
Chen Wei1,2,#, Cai Yicong1,#, Shakeel Ahmad2, Wang Yakun2, An Ruihu2, Tang Shengjia2, Guo Naihui2, Wei Xiangjin2, Tang Shaoqing2, Shao Gaoneng2, Jiao Guiai2, Xie Lihong2, Hu Shikai2, Sheng Zhonghua2( ), Hu Peisong2(
), Hu Peisong2( )
)
Received:2021-09-23
Accepted:2022-01-05
Online:2022-05-28
Published:2022-03-10
Contact:
Sheng Zhonghua, Hu Peisong
About author:First author contact:#These authors contributed equally to this work
Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice[J]. Rice Science, 2022, 29(3): 237-246.
Add to citation manager EndNote|Ris|BibTeX
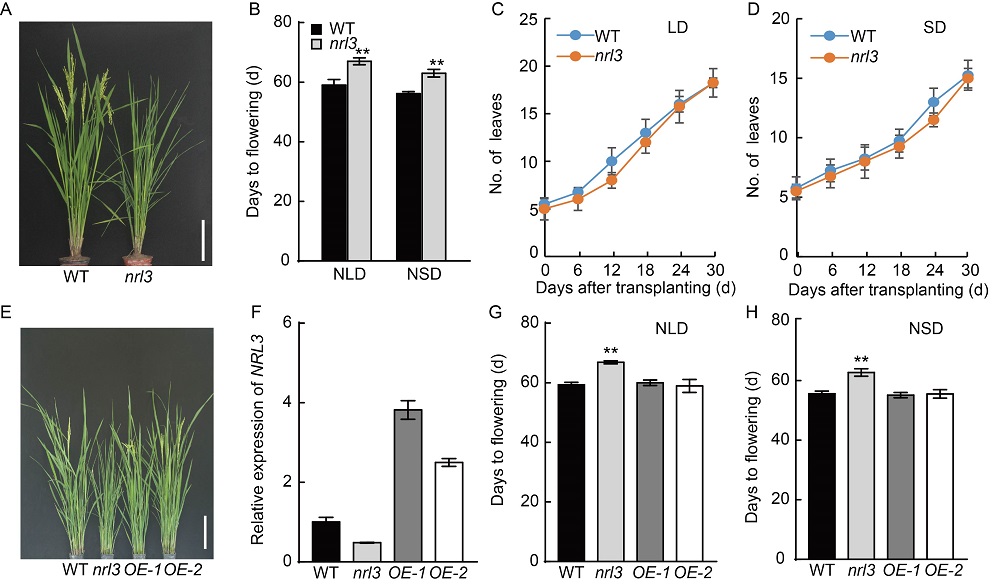
Fig. 1. NRL3 positively regulates heading date under long-day (LD) and short-day (SD) conditions. A, Late heading phenotypes of nrl3 in the field. Scale bar, 20 cm. WT, Wild type (Zhongjiazao 17). B, Heading dates of WT and nrl3 under natural long-day (NLD) and natural short-day (NSD) conditions. Data are Mean ± SD (n = 15). C and D, WT and nrl3 had similar leaf emergence rates under LD (14 h light/10 h dark) and SD (10 h light/14 h dark) conditions. Data are Mean ± SD (n = 10). E, Phenotypes of NRL3 overexpression lines (OE-1 and OE-2) in nrl3 background. Scale bar, 20 cm. F, Expression levels of NRL3 in WT, nrl3 and overexpression lines (OE-1 and OE-2). RNA was extracted from the flag leaves of 60-day-old plants under NLD conditions. The rice Actin gene was used as the internal control. Data are Mean ± SD from three individual replicates. G and H, Heading dates of WT, nrl3 and overexpression lines (OE-1 and OE-2) under NLD and NSD conditions. Data are Mean ± SD (n = 15). Asterisks indicate statistical significance as determined by the Student’s t-test (**, P < 0.01).
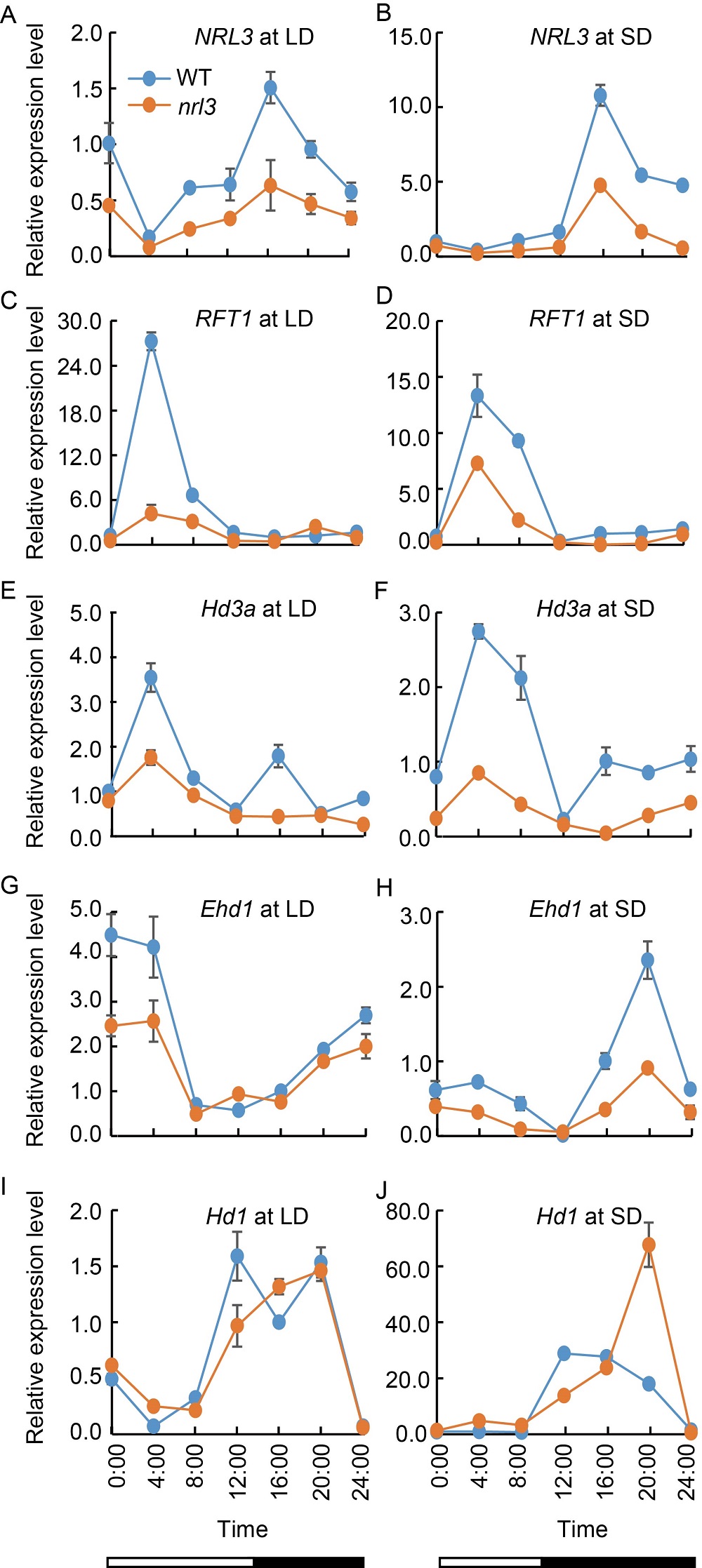
Fig. 2. Expression patterns of NRL3 (A and B), RFT1 (C and D), Hd3a (E and F), Ehd1 (G and H) and Hd1 (I and J) in wild type (WT) Zhongjiazao 17 and nrl3 mutant under long-day (LD) and short-day (SD) conditions. RNA was extracted from the leaves of 20-day-old WT and nrl3 under SD and LD conditions. The rice Actin gene was used as the internal control. Data are Mean ± SD from three individual replicates. Open bars indicate light periods and filled bars indicate dark periods.
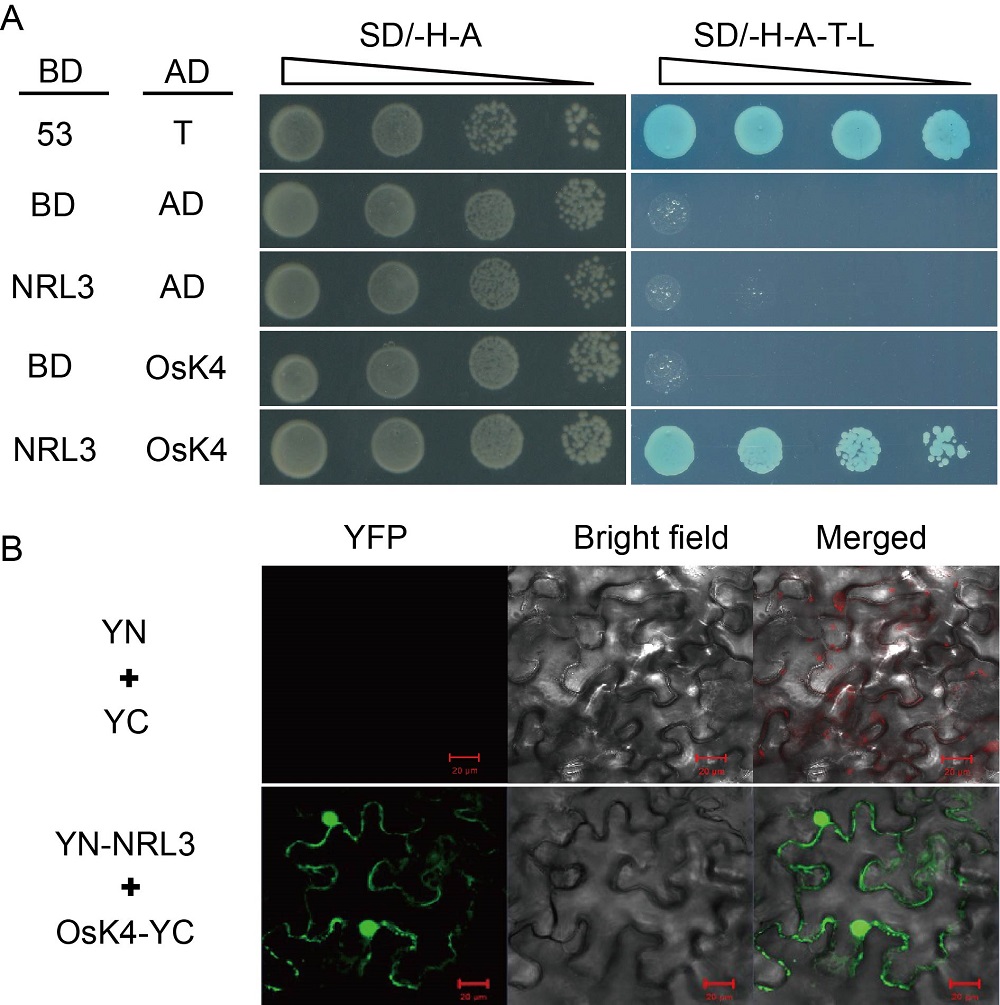
Fig. 3. NRL3 forms a complex with OsK4. A, Interaction between NRL3 and OsK4 in yeast cells. pGBKT7-53 (53) and pGADT7-T (T) were used as positive controls, and pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. SD/-H-A, SD/-Trp-Ade; SD/-H-A-T-L, SD/-His-Ade-Trp-Leu. B, Bimolecular fluorescence complementation (BiFC) analysis of interaction of NRL3 with OsK4 in Nicotiana benthamiana epidermal cells. pSPYNE (YN) and pSPYCE (YC) were used as negative controls. Scale bars, 20 μm. YFP, Yellow fluorescent protein.
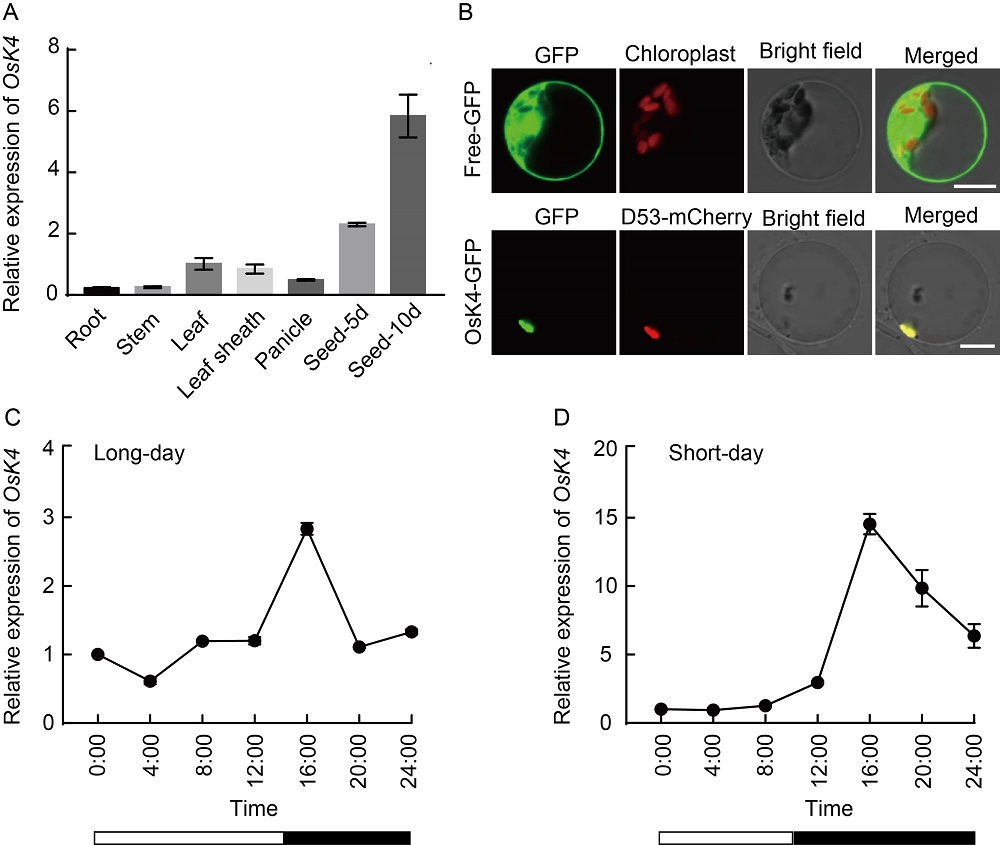
Fig. 4. Expression pattern analysis of OsK4. A, Temporal and spatial expression pattern of OsK4. Seed-5d and seed-10d correspond to developing endosperm at 5 and 10 d after fertilization, respectively. Rice Actin gene was used as the internal control. B, Subcellular location of OsK4-GFP fusion protein in rice protoplasts. Free- GFP (green fluorescent protein) was used as the control, and D53-mCherry as a nuclear marker. Scale bars, 10 μm. C and D, Rhythmic expression patterns of OsK4 under long-day and short-day conditions. Open bars indicate light periods, and filled bars indicate dark periods. RNA was extracted from the leaves of 20-day-old wild type (Zhongjiazao 17) under long-day and short-day conditions. The rice Actin gene was used as an internal control.Data are Mean ± SD from three individual replicates.
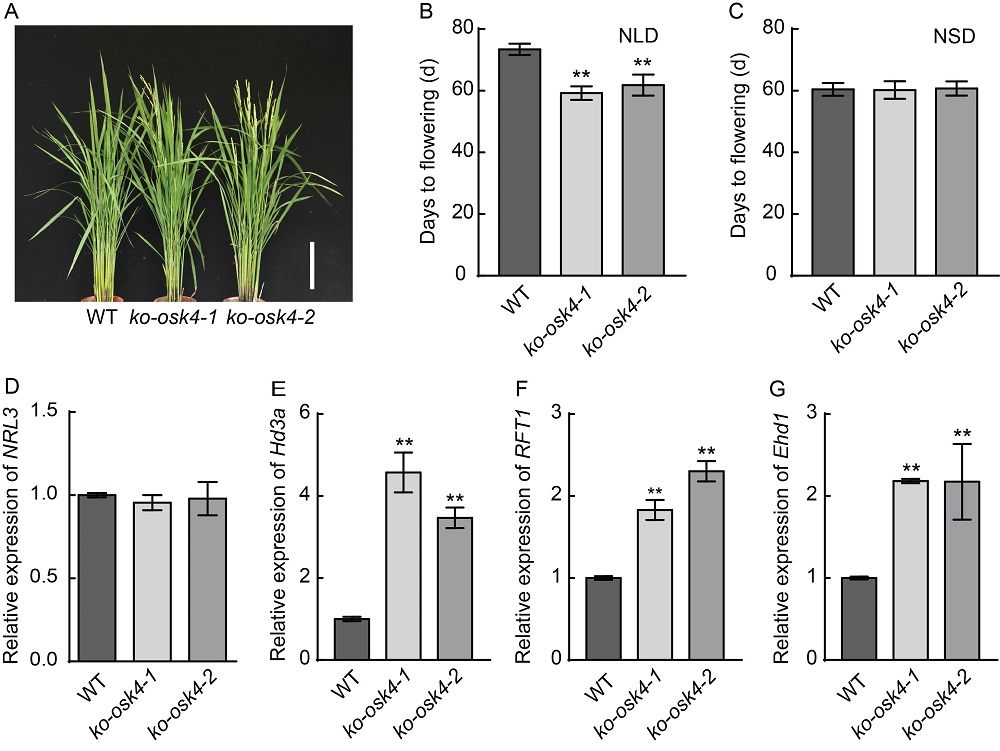
Fig. 5. Phenotypes of OsK4 knock-out transgenic plants and expression patterns of heading date relative genes in ko-osk4 mutants. A, Phenotypes of ko-osk4 plants in field. WT, Wild type (Nipponbare). Scale bar, 20 cm. B and C, Heading date of ko-osk4 plants under natural long-day (NLD) and natural short-day (NSD) conditions. D?G, mRNA levels of NRL3, Hd3a, RFT1 and Ehd1 in WT and ko-osk4. Rice Actin gene was used as the internal control. RNA was extracted from the flag leaves of 60-day-old WT and ko-osk4 mutants under NLD conditions.Data are Mean ± SD from three individual replicates. Asterisks indicated statistical significance as determined by the Student’s t-test (**, P < 0.01).

Fig. 6. Knock-out OsK4 rescues late heading phenotype of nrl3. A, Heading phenotypes of wild type (WT, Zhongjiazao 17), nrl3 mutant and nrl3/osk4 double mutant. Scale bar, 20 cm. Red arrows indicate heading. B, Heading dates of WT, nrl3 and nrl3/osk4 under natural long-day (NLD) conditions. C, Expression levels of OsK4 in WT, nrl3 and nrl3/osk4. RNA was extracted from the leaves of 60-day-old plants under NLD conditions. The rice Actin gene was used as the internal control. D, Expression levels of OsK4 in WT, nrl3 and OE-NRL3. RNA was extracted from the leaves of 60-day-old plants under NLD conditions. The rice Actin gene was used as the internal control. E, Protein levels of OsK4 in WT, nrl3 and nrl3/osk4 by immunoblot analysis. β-ACTIN was used as the internal control. F, OsK4 protein abundance in WT, nrl3 and nrl3/osk4, quantified by ImageJ program. G, Protein levels of OsK4 in WT, nrl3 and OE-NRL3 by immunoblot analysis. β-ACTIN was used as the internal control. H, OsK4 protein abundance in WT, nrl3 and OE-NRL3, quantified by ImageJ program. I, Assay of OsK4 protein stability in vitro. GST-OsK4 proteins were purified from E. coli and incubated with the equal total extracts of WT or nrl3 seedlings in the presence or absence of MG132. GST-OsK4 proteins were detected by GST antibody. J, Quantitative assay of OsK4 protein levels. Data are Mean ± SD from three individual replicates. Asterisks indicate statistical significance as determined by the Student’s t-test (**, P < 0.01).
| [1] | Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, Fornara F. 2017. Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell, 29(11): 2801-2816. |
| [2] | Cai M H, Chen S H, Wu M M, Zheng T H, Zhou L, Li C N, Zhang H, Wang J C, Xu X Y, Chai J T, Ren Y L, Guo X P, Zhang X, Lei C L, Cheng Z J, Wang J, Jiang L, Zhai H Q, Wang H Y, Zhu S S, Wan J M. 2019. Early heading 7 interacts with DTH8, and regulates flowering time in rice. Plant Cell Rep, 38(5): 521-532. |
| [3] | Chai J T, Zhu S S, Li C N, Wang C M, Cai M H, Zheng X M, Zhou L, Zhang H, Sheng P K, Wu M M, Jin X, Cheng Z J, Zhang X, Lei C L, Ren Y L, Lin Q B, Zhou S R, Guo X P, Wang J, Zhao Z C, Wan J M. 2021. OsRE1 interacts with OsRIP1 to regulate rice heading date by finely modulating Ehd1 expression. Plant Biotechnol J, 19(2): 300-310. |
| [4] |
Chen W, Sheng Z H, Cai Y C, Li Q L, Wei X J, Xie L H, Jiao G A, Shao G N, Tang S Q, Wang J L, Hu P S. 2019. Rice morphogenesis and chlorophyll accumulation is regulated by the protein encoded by NRL3 and its interaction with NAL9. Front Plant Sci, 10: 175.
PMID |
| [5] |
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q Z, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science, 316: 1030-1033.
PMID |
| [6] | Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. 2004. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev, 18(8): 926-936. |
| [7] |
Dong H, Fei G L, Wu C Y, Wu F Q, Sun Y Y, Chen M J, Ren Y L, Zhou K N, Cheng Z J, Wang J L, Jiang L, Zhang X, Guo X P, Lei C L, Su N, Wang H Y, Wan J M. 2013. A rice virescent- yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol, 162(4): 1867-1880.
PMID |
| [8] |
Fujino K, Yamanouchi U, Yano M. 2013. Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor Appl Genet, 126(3): 611-618.
PMID |
| [9] | Gao H, Zheng X M, Fei G L, Chen J, Jin M N, Ren Y L, Wu W X, Zhou K N, Sheng P K, Zhou F, Jiang L, Wang J, Zhang X, Guo X P, Wang J L, Cheng Z J, Wu C Y, Wang H Y, Wan J M. 2013. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet, 9(2): e1003281. |
| [10] | Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short- day flowering in rice. Nature, 422: 719-722. |
| [11] | Izawa T. 2007. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot, 58(12): 3091-3097. |
| [12] | Jiang P F, Wang S L, Zheng H, Li H, Zhang F, Su Y H, Xu Z T, Lin H Y, Qian Q, Ding Y. 2018. SIP1 participates in regulation of flowering time in rice by recruiting OsTrx1 to Ehd1. New Phytol, 219(1): 422-435. |
| [13] | Jung C, Müller A E. 2009. Flowering time control and applications in plant breeding. Trends Plant Sci, 14(10): 563-573. |
| [14] | Kim S L, Lee S, Kim H J, Nam H G, An G. 2007. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol, 145(4): 1484-1494. |
| [15] |
Komiya R, Yokoi S, Shimamoto K. 2009. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development, 136(20): 3443-3450.
PMID |
| [16] | Lee Y S, Jeong D H, Lee D Y, Yi J, Ryu C H, Kim S L, Jeong H J, Choi S C, Jin P, Yang J, Cho L H, Choi H, An G. 2010. OsCOL4is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J, 63(1): 18-30. |
| [17] | Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez J P, Eshed Y. 2006. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA, 103: 6398-6403. |
| [18] | Liu K P, Yu Y, Dong A W, Shen W H. 2017. SET DOMAIN GROUP701 encodes a H3K4-methytransferase and regulates multiple key processes of rice plant development. New Phytol, 215(2): 609-623. |
| [19] | Liu X F, Li M, Liu K, Tang D, Sun M F, Li Y F, Shen Y, Du G J, Cheng Z K. 2016. Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J Exp Bot, 67(8): 2139-2150. |
| [20] |
Ma L, Sang X C, Zhang T, Yu Z Y, Li Y F, Zhao F M, Wang Z W, Wang Y T, Yu P, Wang N, Zhang C W, Ling Y H, Yang Z L, He G H. 2017. ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol, 213(1): 275-286.
PMID |
| [21] | Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang Z X, Minobe Y, Yano M. 2011. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J, 66(4): 603-612. |
| [22] | Nemoto Y, Nonoue Y, Yano M, Izawa T. 2016. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J, 86(3): 221-233. |
| [23] |
Onouchi H, Igeño M I, Périlleux C, Graves K, Coupland G. 2000. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell, 12(6): 885-900.
PMID |
| [24] | Peng L T, Shi Z Y, Li L, Shen G Z, Zhang J L. 2008. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J Plant Physiol, 165(8): 876-885. |
| [25] | Samach A, Onouchi H, Gold S E, Ditta G S, Schwarz-Sommer Z, Yanofsky M F, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science, 2888: 1613-1616. |
| [26] | Sheng P K, Wu F Q, Tan J J, Zhang H, Ma W W, Chen L P, Wang J C, Wang J, Zhu S S, Guo X P, Wang J L, Zhang X, Cheng Z J, Bao Y Q, Wu C Y, Liu X M, Wan J M. 2016. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol Biol, 92(1/2): 209-222. |
| [27] |
Simpson G G. 2003. Evolution of flowering in response to day length: Flipping the CONSTANS switch. BioEssays, 25(9): 829-832.
PMID |
| [28] |
Song S, Wang G F, Hu Y, Liu H Y, Bai X F, Qin R, Xing Y Z. 2018. OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J Exp Bot, 69(18): 4283-4293.
PMID |
| [29] | Sun C H, Chen D, Fang J, Wang P R, Deng X J, Chu C C. 2014. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell, 5(12): 889-898. |
| [30] | Sun X H, Zhang Z G, Wu J X, Cui X A, Feng D, Wang K, Xu M, Zhou L, Han X, Gu X F, Lu T G. 2016. The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLoS Genet, 12(3): e1005927. |
| [31] | Tan J J, Jin M N, Wang J C, Wu F Q, Sheng P K, Cheng Z J, Wang J L, Zheng X M, Chen L P, Wang M, Zhu S S, Guo X P, Zhang X, Liu X M, Wang C M, Wang H Y, Wu C Y, Wan J M. 2016. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol, 57(4): 798-812. |
| [32] | Tsuji H, Taoka K I, Shimamoto K. 2011. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol, 14(1): 45-52. |
| [33] | Turck F, Fornara F, Coupland G. 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol, 59: 573-594. |
| [34] | Wang F, Zhu D M, Huang X, Li S, Gong Y N, Yao Q F, Fu X D, Fan L M, Deng X W. 2009. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell, 21: 2378-2390. |
| [35] | Wang P, Gong R, Yang Y, Yu S B. 2019. Ghd8 controls rice photoperiod sensitivity by forming a complex that interacts with Ghd7. BMC Plant Biol, 19(1): 462. |
| [36] | Wei X J, Xu J F, Guo H N, Jiang L, Chen S H, Yu C Y, Zhou Z L, Hu P S, Zhai H Q, Wan J M. 2010. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol, 153(4): 1747-1758. |
| [37] | Yang Y, Fu D B, Zhu C M, He Y Z, Zhang H J, Liu T, Li X H, Wu C Y. 2015. The RING-finger ubiquitin ligase HAF1 mediates heading date 1 degradation during photoperiodic flowering in rice. Plant Cell, 27(9): 2455-2468. |
| [38] |
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell, 12(12): 2473-2484.
PMID |
| [39] |
Zhang Y, Su J B, Duan S, Ao Y, Dai J R, Liu J, Wang P, Li Y G, Liu B, Feng D R, Wang J F, Wang H B. 2011. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods, 7(1): 30.
PMID |
| [40] | Zhang Z Y, Hu W, Shen G J, Liu H Y, Hu Y, Zhou X C, Liu T M, Xing Y Z. 2017. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep, 7(1): 5388. |
| [41] | Zhao S S, Zhao L, Liu F X, Wu Y Z, Zhu Z F, Sun C Q, Tan L B. 2016. NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J Integr Plant Biol, 58(12): 983-996. |
| [42] | Zhu S S, Wang J C, Cai M H, Zhang H, Wu F Q, Xu Y, Li C N, Cheng Z J, Zhang X, Guo X P, Sheng P K, Wu M M, Wang J L, Lei C L, Wang J, Zhao Z C, Wu C Y, Wang H Y, Wan J M. 2017. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J Exp Bot, 68(3): 553-568. |
| [1] | Md. Dhin Islam, Adam H. Price, Paul D. Hallett. Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth [J]. Rice Science, 2023, 30(5): 459-472. |
| [2] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [3] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [4] | Shuting Yuan, Chunjue Xu, Wei Yan, Zhenyi Chang, Xingwang Deng, Zhufeng Chen, Jianxin Wu, Xiaoyan Tang. Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis [J]. Rice Science, 2020, 27(4): 289-301. |
| [5] | Vijayaraghavareddy Preethi, Xinyou Yin, C. Struik Paul, Makarla Udayakumar, Sreeman Sheshshayee. Responses of Lowland, Upland and Aerobic Rice Genotypes to Water Limitation During Different Phases [J]. Rice Science, 2020, 27(4): 345-354. |
| [6] | Hussain Kashif, Yingxing Zhang, Anley Workie, Riaz Aamir, Abbas Adil, Hasanuzzaman Rani Md., Hong Wang, Xihong Shen, Liyong Cao, Shihua Cheng. Association Mapping of Quantitative Trait Loci for Grain Size in Introgression Line Derived from Oryza rufipogon [J]. Rice Science, 2020, 27(3): 246-254. |
| [7] | Hua Zhang, Xu Liu, Yongyi Yang, Ning Xuan, Fangyin Yao. Mapping of Hd-6-2 for Heading Date Using Two Secondary Segregation Populations in Rice [J]. Rice Science, 2018, 25(3): 161-168. |
| [8] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [9] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| [10] | Fernando Polesi Luís, Bruder Silveira Sarmento Silene, Guidolin Canniatti-Brazaca Solange. Starch Digestibility and Functional Properties of Rice Starch Subjected to Gamma Radiation [J]. Rice Science, 2018, 25(1): 42-51. |
| [11] | Singh Bhupinder, Raja Reddy Kambham, Diaz Redoña Edilberto, Walker Timothy. Screening of Rice Cultivars for Morpho-Physiological Responses to Early-Season Soil Moisture Stress [J]. Rice Science, 2017, 24(6): 322-335. |
| [12] | Jini D., Joseph B.. Physiological Mechanism of Salicylic Acid for Alleviation of Salt Stress in Rice [J]. Rice Science, 2017, 24(2): 97-108. |
| [13] | Fernando Polesi Luís, Divino da Matta Junior Manoel, Bruder Silveira Sarmento Silene, Guidolin Canniatti-Brazaca Solange. Starch Digestibility and Physicochemical and Cooking Properties of Irradiated Rice Grains [J]. Rice Science, 2017, 24(1): 48-55. |
| [14] | S. Abe S., Yamasaki Y., Wakatsuki T.. Assessing Silicon Availability in Soils of Rice-Growing Lowlands and Neighboring Uplands in Benin and Nigeria [J]. Rice Science, 2016, 23(4): 196-202. |
| [15] | Saleethong Paweena, Roytrakul Sittiruk, Kong-Ngern Kanlaya, Theerakulpisut Piyada. Differential Proteins Expressed in Rice Leaves and Grains in Response to Salinity and Exogenous Spermidine Treatments [J]. Rice Science, 2016, 23(1): 9-21. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||