
Rice Science ›› 2021, Vol. 28 ›› Issue (4): 379-390.DOI: 10.1016/j.rsci.2021.05.008
• Research Papers • Previous Articles Next Articles
Rakotoson Tatiana1,3, Dusserre Julie4,7, Letourmy Philippe4,7, Frouin Julien5,7, Ramonta Ratsimiala Isabelle2, Victorine Rakotoarisoa Noronirina2, Cao Tuong-Vi5,7, Vom Brocke Kirsten1,6,7, Ramanantsoanirina Alain1, Ahmadi Nourollah5,7, Raboin Louis-Marie4,7
Received:2020-04-04
Accepted:2020-11-02
Online:2021-07-28
Published:2021-07-28
Rakotoson Tatiana, Dusserre Julie, Letourmy Philippe, Frouin Julien, Ramonta Ratsimiala Isabelle, Victorine Rakotoarisoa Noronirina, Cao Tuong-Vi, Vom Brocke Kirsten, Ramanantsoanirina Alain, Ahmadi Nourollah, Raboin Louis-Marie. Genome-Wide Association Study of Nitrogen Use Efficiency and Agronomic Traits in Upland Rice[J]. Rice Science, 2021, 28(4): 379-390.
Add to citation manager EndNote|Ris|BibTeX
| Code | Trait | Formulas | Units |
|---|---|---|---|
| AL | Awn length | Length of awn using a scale of 1 (no awn) to 9 (very long awn) | - |
| DTF | Days to flowering | Days from sowing to flowering | days |
| FG | Filled grain | 100 × number of filled spikelets /total number of spikelets | % |
| GNC | Grain N content | Grain N concentration of 6 hills at maturity | % |
| GL | Grain Length | Length of grain | mm |
| GP | Grain Pilosity | Pilosity of grain using a scale of 1 (glabrous) to 9 (very hairy) | - |
| GT | Grain Thickness | Thickness of grain | mm |
| GW | Grain Width | Width of grain | mm |
| GY | Grain yield | PANM2 × SPIPAN × FG × TGW × 10 | kg/hm2 |
| HI | Harvest index | GY / (GY + SY) | - |
| NHI | Nitrogen harvest index | GNC × GY / TNUP x 100 | - |
| NUE | Nitrogen use efficiency | GY / N supply | kg/kg |
| NUPE | Nitrogen uptake efficiency | TNUP / N supply | kg/kg |
| NUTE | Nitrogen utilization efficiency | GY / TNUP | kg/kg |
| PANM2 | Number of panicles per m² | Number of panicle of 6 hills / 0.24 | - |
| PH | Plant height | Height of plant from soil surface to the tip of main panicle | cm |
| SNC | Straw N content | Straw N concentration of 6 hills at maturity | % |
| SPIPAN | Number of spikelets per panicle | Number of spikelets of 6 hills / number of panicle of 6 hills | - |
| SY | Straw yield | 10 × Biomass of 6 hills / 0.24 | kg/hm2 |
| TGW | 1000-grain weight | Weight of 200 filled spikelets × 5 | g |
| TNUP | Total plant N uptake | (GY × GNC/100) + (SY × SNC/100) | kg/hm2 |
Table S1. Description of 21 measured and calculated traits.
| Code | Trait | Formulas | Units |
|---|---|---|---|
| AL | Awn length | Length of awn using a scale of 1 (no awn) to 9 (very long awn) | - |
| DTF | Days to flowering | Days from sowing to flowering | days |
| FG | Filled grain | 100 × number of filled spikelets /total number of spikelets | % |
| GNC | Grain N content | Grain N concentration of 6 hills at maturity | % |
| GL | Grain Length | Length of grain | mm |
| GP | Grain Pilosity | Pilosity of grain using a scale of 1 (glabrous) to 9 (very hairy) | - |
| GT | Grain Thickness | Thickness of grain | mm |
| GW | Grain Width | Width of grain | mm |
| GY | Grain yield | PANM2 × SPIPAN × FG × TGW × 10 | kg/hm2 |
| HI | Harvest index | GY / (GY + SY) | - |
| NHI | Nitrogen harvest index | GNC × GY / TNUP x 100 | - |
| NUE | Nitrogen use efficiency | GY / N supply | kg/kg |
| NUPE | Nitrogen uptake efficiency | TNUP / N supply | kg/kg |
| NUTE | Nitrogen utilization efficiency | GY / TNUP | kg/kg |
| PANM2 | Number of panicles per m² | Number of panicle of 6 hills / 0.24 | - |
| PH | Plant height | Height of plant from soil surface to the tip of main panicle | cm |
| SNC | Straw N content | Straw N concentration of 6 hills at maturity | % |
| SPIPAN | Number of spikelets per panicle | Number of spikelets of 6 hills / number of panicle of 6 hills | - |
| SY | Straw yield | 10 × Biomass of 6 hills / 0.24 | kg/hm2 |
| TGW | 1000-grain weight | Weight of 200 filled spikelets × 5 | g |
| TNUP | Total plant N uptake | (GY × GNC/100) + (SY × SNC/100) | kg/hm2 |
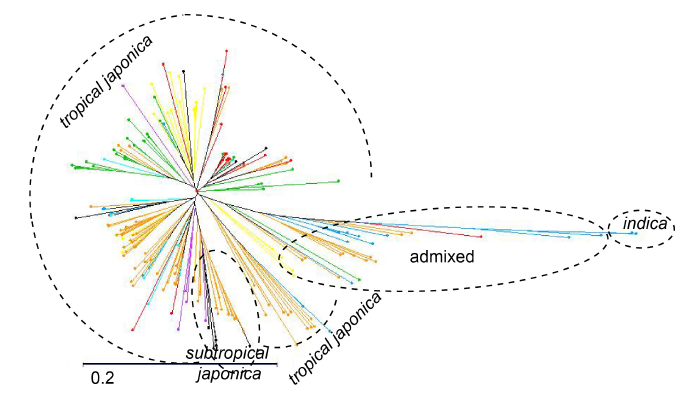
Fig. 1. Neighbour joining tree of the 190 accessions based on 38 390 single nucleotide polymorphisms (SNPs). Accessions are coloured according to the breeding center from which they originate. FOFIFA (National Center for Applied Research on Rural Development)/CIRAD (French Agricultural Research Centre for International Development) in Madagascar (Orange); CIAT (International Center for Tropical Agriculture) in Colombia (Yellow); EMBRAPA (Brazilian Agricultural Research Corporation) in Brazil (Red); CIRAD in Brazil (Blue); CIRAD in Ivory coast (Light blue); Africa rice in Ivory coast (Green); Yunnan Academy of Agricultural Sciences in China (Violet).
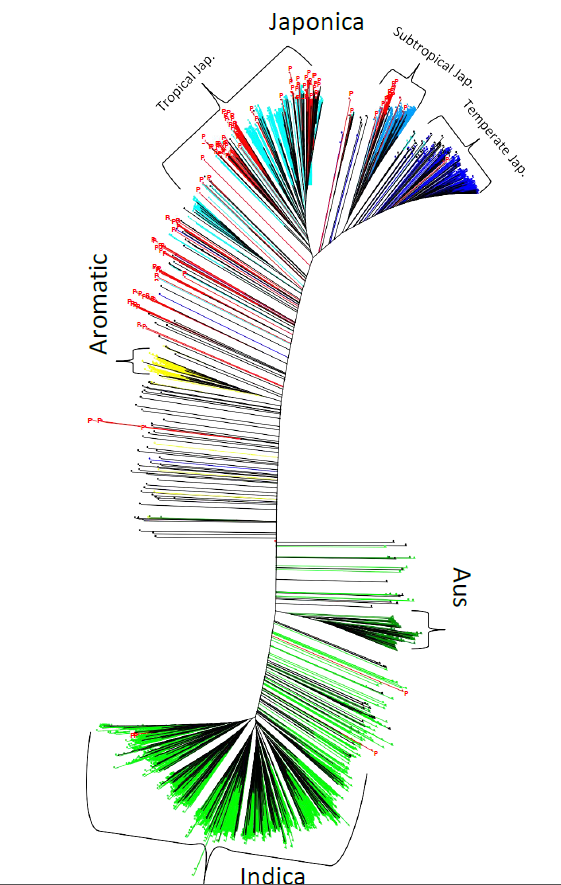
Fig. S1. Neighbour-joining tree based on 27 326 genotyping-by-sequencing single nucleotide polymorphisms.Accessions from the panel are coloured red and tagged with the letter «P».
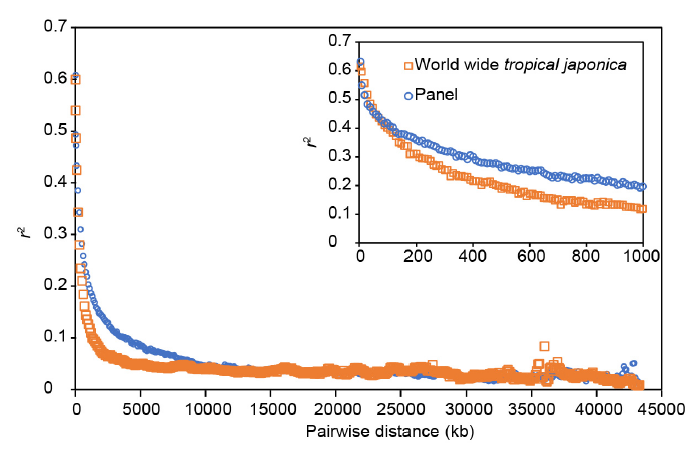
Fig. 2. Comparison of linkage disequilibrium (LD) decay in panel of 190 accessions used in this study (in red) and the worldwide tropical japonica reference panel (in blue). The curves represent the average r² among the 12 chromosomes over the whole length of the chromosome (main frame) or in 10 kb bins for LD decay plot over a maximum distance of 1000 kb (zoomed frame). Each circle corresponds to the average LD between marker pairs in 100 kb bins.
| Chr. | SNP position | Trait | Max | Min | LOD | R2 (%) | Qmin | Candidate gene within 100 kb |
|---|---|---|---|---|---|---|---|---|
| 1 | 594 602 | GNC | 179 | 11 | 4.22 | 9.0 | 0.68 | OsNRT2.1 |
| 1 | 594 602 | GNC | 179 | 11 | 4.22 | 9.0 | 0.68 | OsNIP4.1 |
| 1 | 2 961 416 | FG, DTF, NHI, HI | 172 | 18 | 4.97 | 10.3 | 0.03 | OsCrRLK1L4 |
| 1 | 2 972 214 | NHI, DTF, HI | 171 | 19 | 5.09 | 10.6 | 0.03 | |
| 1 | 2 972 304 | DTF, NHI | 178 | 12 | 4.77 | 8.8 | 0.13 | |
| 1 | 3 028 472 | DTF, SY, NHI, HI | 177 | 13 | 5.25 | 10.4 | 0.03 | |
| 1 | 3 031 975 | NUTE, NHI, HI | 178 | 12 | 5.51 | 11.0 | 0.03 | |
| 1 | 3 034 970 | NUTE, NHI, HI | 181 | 9 | 6.00 | 11.8 | 0.02 | |
| 1 | 4 825 760 | NUTE, NHI, HI | 181 | 9 | 7.26 | 14.9 | 0.00 | |
| 1 | 4 958 939 | NHI | 173 | 17 | 4.07 | 7.5 | 0.17 | OsDOS |
| 1 | 7 309 207 | SNC | 104 | 86 | 4.09 | 8.7 | 0.55 | OsTIP4;2 |
| 1 | 29 135 405 | GNC, NUTE | 179 | 11 | 4.76 | 10.4 | 0.13 | OsNRT2.3a |
| 1 | 29 212 814 | GNC | 177 | 13 | 4.01 | 8.5 | 0.13 | CBSCLC2 |
| 1 | 33 331 630 | SY | 155 | 35 | 4.23 | 8.8 | 0.47 | OsGH3.1 |
| 1 | 35 227 489 | TNUP | 141 | 49 | 4.03 | 8.2 | 0.73 | OsAAT7 |
| 2 | 2 843 055 | DTF | 136 | 54 | 4.19 | 9.0 | 0.28 | lc2; OsVIL2 |
| 2 | 24 480 078 | TGW | 155 | 35 | 4.17 | 7.3 | 0.19 | OsMPS |
| 2 | 24 510 877 | TGW | 121 | 69 | 4.09 | 8.1 | 0.19 | |
| 3 | 1 352 456 | DTF | 157 | 33 | 8.15 | 19.9 | 0.00 | DTH3 |
| 3 | 1 356 326 | DTF | 164 | 26 | 8.12 | 19.9 | 0.00 | |
| 3 | 1 395 165 | DTF, FG, NUTE | 160 | 30 | 7.01 | 16.7 | 0.00 | OsCrRLK1L6 |
| 3 | 1 424 348 | DTF | 154 | 36 | 8.81 | 21.9 | 0.00 | |
| 3 | 3 425 919 | PH | 167 | 23 | 4.88 | 6.7 | 0.20 | oscow1; nal7 |
| 3 | 3 602 444 | PH | 175 | 15 | 4.27 | 5.7 | 0.20 | |
| 3 | 3 677 867 | PH | 174 | 16 | 4.23 | 5.6 | 0.23 | |
| 3 | 3 728 449 | PH | 175 | 15 | 4.27 | 5.7 | 0.20 | OsDof12 |
| 3 | 3 845 920 | DTF | 176 | 14 | 5.34 | 12.1 | 0.03 | OsSUT1 |
| 3 | 16 174 183 | TGW, GL | 135 | 55 | 5.46 | 11.8 | 0.03 | GS3 |
| 3 | 16 174 184 | TGW, GL | 135 | 55 | 5.46 | 11.8 | 0.03 | |
| 3 | 16 414 381 | TGW | 147 | 43 | 4.23 | 8.4 | 0.20 | |
| 3 | 16 790 082 | TGW, GL | 152 | 38 | 6.17 | 13.6 | 0.02 | |
| 4 | 31 050 939 | PANM2 | 155 | 35 | 5.85 | 10.1 | 0.02 | osks1 |
| 4 | 31 070 568 | PANM2 | 172 | 18 | 4.69 | 8.3 | 0.05 | osks2 |
| 4 | 31 243 064 | PANM2 | 144 | 46 | 4.54 | 9.6 | 0.09 | 15 masked associations with PANM2 |
| 4 | 31 250 082 | PANM2 | 141 | 49 | 5.92 | 10.8 | 0.02 | nal1, GPS, SPIKE, LSCHL4 |
| 4 | 31 450 557 | PANM2, PH | 159 | 31 | 5.70 | 11.6 | 0.03 | Six masked associations with PANM2 |
| 5 | 3 506 138 | GW | 157 | 33 | 5.09 | 9.2 | 0.10 | GS5 |
| 5 | 5 309 762 | GW | 158 | 32 | 4.05 | 6.8 | 0.18 | GW5 |
| 5 | 5 361 979 | GW | 113 | 77 | 4.55 | 7.7 | 0.07 | |
| 5 | 5 362 675 | GW | 112 | 78 | 6.01 | 10.7 | 0.01 | |
| 5 | 5 362 698 | GW | 114 | 76 | 6.08 | 11.1 | 0.01 | |
| 5 | 5 365 520 | GW | 114 | 76 | 4.02 | 7.0 | 0.18 | |
| 5 | 5 391 748 | GW | 122 | 68 | 6.48 | 11.7 | 0.01 | Five masked associations with PANM2 |
| 6 | 21 981 979 | TNUP | 157 | 33 | 4.15 | 8.7 | 0.38 | d35 |
| 6 | 21 982 001 | TNUP | 157 | 33 | 4.15 | 8.7 | 0.38 | |
| 6 | 22 028 211 | TNUP | 161 | 29 | 4.10 | 8.5 | 0.38 | |
| 6 | 22 047 685 | TNUP | 161 | 29 | 4.10 | 8.5 | 0.38 | |
| 6 | 22 089 331 | TNUP, NUPE | 162 | 28 | 4.48 | 9.5 | 0.38 | |
| 9 | 1 504 274 | GL | 169 | 21 | 4.19 | 8.6 | 0.15 | |
| 9 | 1 504 282 | GL | 169 | 21 | 4.19 | 8.6 | 0.15 | |
| 9 | 1 824 735 | NUTE | 169 | 21 | 4.10 | 8.4 | 0.08 | rFCA |
| 11 | 19 200 467 | GY, NUE, NUTE, NHI, HI | 180 | 10 | 5.53 | 11.6 | 0.03 | tld1-D |
| 11 | 19 365 597 | NUTE, NHI, HI | 181 | 9 | 4.72 | 8.7 | 0.13 | |
| 12 | 9 914 173 | NHI | 126 | 64 | 4.46 | 8.3 | 0.14 | rbcS, OsRBCS2 |
| 12 | 10 531 085 | NHI | 126 | 64 | 4.84 | 9.2 | 0.13 |
Table 1 Results of genome-wide association analysis for SNPs from haplotypes close to relevant putative genes.
| Chr. | SNP position | Trait | Max | Min | LOD | R2 (%) | Qmin | Candidate gene within 100 kb |
|---|---|---|---|---|---|---|---|---|
| 1 | 594 602 | GNC | 179 | 11 | 4.22 | 9.0 | 0.68 | OsNRT2.1 |
| 1 | 594 602 | GNC | 179 | 11 | 4.22 | 9.0 | 0.68 | OsNIP4.1 |
| 1 | 2 961 416 | FG, DTF, NHI, HI | 172 | 18 | 4.97 | 10.3 | 0.03 | OsCrRLK1L4 |
| 1 | 2 972 214 | NHI, DTF, HI | 171 | 19 | 5.09 | 10.6 | 0.03 | |
| 1 | 2 972 304 | DTF, NHI | 178 | 12 | 4.77 | 8.8 | 0.13 | |
| 1 | 3 028 472 | DTF, SY, NHI, HI | 177 | 13 | 5.25 | 10.4 | 0.03 | |
| 1 | 3 031 975 | NUTE, NHI, HI | 178 | 12 | 5.51 | 11.0 | 0.03 | |
| 1 | 3 034 970 | NUTE, NHI, HI | 181 | 9 | 6.00 | 11.8 | 0.02 | |
| 1 | 4 825 760 | NUTE, NHI, HI | 181 | 9 | 7.26 | 14.9 | 0.00 | |
| 1 | 4 958 939 | NHI | 173 | 17 | 4.07 | 7.5 | 0.17 | OsDOS |
| 1 | 7 309 207 | SNC | 104 | 86 | 4.09 | 8.7 | 0.55 | OsTIP4;2 |
| 1 | 29 135 405 | GNC, NUTE | 179 | 11 | 4.76 | 10.4 | 0.13 | OsNRT2.3a |
| 1 | 29 212 814 | GNC | 177 | 13 | 4.01 | 8.5 | 0.13 | CBSCLC2 |
| 1 | 33 331 630 | SY | 155 | 35 | 4.23 | 8.8 | 0.47 | OsGH3.1 |
| 1 | 35 227 489 | TNUP | 141 | 49 | 4.03 | 8.2 | 0.73 | OsAAT7 |
| 2 | 2 843 055 | DTF | 136 | 54 | 4.19 | 9.0 | 0.28 | lc2; OsVIL2 |
| 2 | 24 480 078 | TGW | 155 | 35 | 4.17 | 7.3 | 0.19 | OsMPS |
| 2 | 24 510 877 | TGW | 121 | 69 | 4.09 | 8.1 | 0.19 | |
| 3 | 1 352 456 | DTF | 157 | 33 | 8.15 | 19.9 | 0.00 | DTH3 |
| 3 | 1 356 326 | DTF | 164 | 26 | 8.12 | 19.9 | 0.00 | |
| 3 | 1 395 165 | DTF, FG, NUTE | 160 | 30 | 7.01 | 16.7 | 0.00 | OsCrRLK1L6 |
| 3 | 1 424 348 | DTF | 154 | 36 | 8.81 | 21.9 | 0.00 | |
| 3 | 3 425 919 | PH | 167 | 23 | 4.88 | 6.7 | 0.20 | oscow1; nal7 |
| 3 | 3 602 444 | PH | 175 | 15 | 4.27 | 5.7 | 0.20 | |
| 3 | 3 677 867 | PH | 174 | 16 | 4.23 | 5.6 | 0.23 | |
| 3 | 3 728 449 | PH | 175 | 15 | 4.27 | 5.7 | 0.20 | OsDof12 |
| 3 | 3 845 920 | DTF | 176 | 14 | 5.34 | 12.1 | 0.03 | OsSUT1 |
| 3 | 16 174 183 | TGW, GL | 135 | 55 | 5.46 | 11.8 | 0.03 | GS3 |
| 3 | 16 174 184 | TGW, GL | 135 | 55 | 5.46 | 11.8 | 0.03 | |
| 3 | 16 414 381 | TGW | 147 | 43 | 4.23 | 8.4 | 0.20 | |
| 3 | 16 790 082 | TGW, GL | 152 | 38 | 6.17 | 13.6 | 0.02 | |
| 4 | 31 050 939 | PANM2 | 155 | 35 | 5.85 | 10.1 | 0.02 | osks1 |
| 4 | 31 070 568 | PANM2 | 172 | 18 | 4.69 | 8.3 | 0.05 | osks2 |
| 4 | 31 243 064 | PANM2 | 144 | 46 | 4.54 | 9.6 | 0.09 | 15 masked associations with PANM2 |
| 4 | 31 250 082 | PANM2 | 141 | 49 | 5.92 | 10.8 | 0.02 | nal1, GPS, SPIKE, LSCHL4 |
| 4 | 31 450 557 | PANM2, PH | 159 | 31 | 5.70 | 11.6 | 0.03 | Six masked associations with PANM2 |
| 5 | 3 506 138 | GW | 157 | 33 | 5.09 | 9.2 | 0.10 | GS5 |
| 5 | 5 309 762 | GW | 158 | 32 | 4.05 | 6.8 | 0.18 | GW5 |
| 5 | 5 361 979 | GW | 113 | 77 | 4.55 | 7.7 | 0.07 | |
| 5 | 5 362 675 | GW | 112 | 78 | 6.01 | 10.7 | 0.01 | |
| 5 | 5 362 698 | GW | 114 | 76 | 6.08 | 11.1 | 0.01 | |
| 5 | 5 365 520 | GW | 114 | 76 | 4.02 | 7.0 | 0.18 | |
| 5 | 5 391 748 | GW | 122 | 68 | 6.48 | 11.7 | 0.01 | Five masked associations with PANM2 |
| 6 | 21 981 979 | TNUP | 157 | 33 | 4.15 | 8.7 | 0.38 | d35 |
| 6 | 21 982 001 | TNUP | 157 | 33 | 4.15 | 8.7 | 0.38 | |
| 6 | 22 028 211 | TNUP | 161 | 29 | 4.10 | 8.5 | 0.38 | |
| 6 | 22 047 685 | TNUP | 161 | 29 | 4.10 | 8.5 | 0.38 | |
| 6 | 22 089 331 | TNUP, NUPE | 162 | 28 | 4.48 | 9.5 | 0.38 | |
| 9 | 1 504 274 | GL | 169 | 21 | 4.19 | 8.6 | 0.15 | |
| 9 | 1 504 282 | GL | 169 | 21 | 4.19 | 8.6 | 0.15 | |
| 9 | 1 824 735 | NUTE | 169 | 21 | 4.10 | 8.4 | 0.08 | rFCA |
| 11 | 19 200 467 | GY, NUE, NUTE, NHI, HI | 180 | 10 | 5.53 | 11.6 | 0.03 | tld1-D |
| 11 | 19 365 597 | NUTE, NHI, HI | 181 | 9 | 4.72 | 8.7 | 0.13 | |
| 12 | 9 914 173 | NHI | 126 | 64 | 4.46 | 8.3 | 0.14 | rbcS, OsRBCS2 |
| 12 | 10 531 085 | NHI | 126 | 64 | 4.84 | 9.2 | 0.13 |
| Parameter | Haplotype | GY | PANM2 | SPIPAN | FG | TGW | DTF | GL | GW | GT | PH | SY | HI | NUE | NUPE | NUTE | SNC | GNC | TNUP | NHI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | 57 | 3 | 18 | 26 | 12 | 28 | 27 | 10 | 24 | 9 | 49 | 4 | 5 | 28 | 2 | 26 | 10 | 31 | ||
| GY | 1 | 3 | 1 | 1 | ||||||||||||||||
| PANM2 | 3 | 1 | 3 | |||||||||||||||||
| SPIPAN | 3 | 1 | ||||||||||||||||||
| FG | 7 | 2 | 1 | 2 | 1 | 1 | 1 | |||||||||||||
| TGW | 7 | 1 | 3 | |||||||||||||||||
| DTF | 4 | 4 | 1 | 4 | 1 | 5 | ||||||||||||||
| GL | 4 | 2 | ||||||||||||||||||
| GW | 6 | 1 | 1 | 1 | ||||||||||||||||
| GT | 2 | 1 | ||||||||||||||||||
| PH | 8 | 1 | 2 | 1 | 1 | 1 | ||||||||||||||
| SY | 4 | 2 | 1 | 1 | 1 | 1 | ||||||||||||||
| HI | 12 | 2 | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 1 | 6 | 21 | ||||||||
| NUE | 2 | 2 | 1 | 2 | 1 | 1 | ||||||||||||||
| NUPE | 2 | 1 | 1 | 1 | 1 | 5 | ||||||||||||||
| NUTE | 10 | 2 | 1 | 4 | 3 | 1 | 1 | 4 | 2 | 1 | 13 | 6 | ||||||||
| SNC | 1 | 1 | ||||||||||||||||||
| GNC | 5 | 1 | 1 | 1 | 1 | 1 | 3 | |||||||||||||
| TNUP | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |||||||||||
| NHI | 9 | 1 | 2 | 2 | 2 | 1 | 6 | 1 | 4 | 1 |
Table 2 Results of counting significant SNPs or haplotypes shared by traits.
| Parameter | Haplotype | GY | PANM2 | SPIPAN | FG | TGW | DTF | GL | GW | GT | PH | SY | HI | NUE | NUPE | NUTE | SNC | GNC | TNUP | NHI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | 57 | 3 | 18 | 26 | 12 | 28 | 27 | 10 | 24 | 9 | 49 | 4 | 5 | 28 | 2 | 26 | 10 | 31 | ||
| GY | 1 | 3 | 1 | 1 | ||||||||||||||||
| PANM2 | 3 | 1 | 3 | |||||||||||||||||
| SPIPAN | 3 | 1 | ||||||||||||||||||
| FG | 7 | 2 | 1 | 2 | 1 | 1 | 1 | |||||||||||||
| TGW | 7 | 1 | 3 | |||||||||||||||||
| DTF | 4 | 4 | 1 | 4 | 1 | 5 | ||||||||||||||
| GL | 4 | 2 | ||||||||||||||||||
| GW | 6 | 1 | 1 | 1 | ||||||||||||||||
| GT | 2 | 1 | ||||||||||||||||||
| PH | 8 | 1 | 2 | 1 | 1 | 1 | ||||||||||||||
| SY | 4 | 2 | 1 | 1 | 1 | 1 | ||||||||||||||
| HI | 12 | 2 | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 1 | 6 | 21 | ||||||||
| NUE | 2 | 2 | 1 | 2 | 1 | 1 | ||||||||||||||
| NUPE | 2 | 1 | 1 | 1 | 1 | 5 | ||||||||||||||
| NUTE | 10 | 2 | 1 | 4 | 3 | 1 | 1 | 4 | 2 | 1 | 13 | 6 | ||||||||
| SNC | 1 | 1 | ||||||||||||||||||
| GNC | 5 | 1 | 1 | 1 | 1 | 1 | 3 | |||||||||||||
| TNUP | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |||||||||||
| NHI | 9 | 1 | 2 | 2 | 2 | 1 | 6 | 1 | 4 | 1 |
| Genotype | Country | Breeding center | Panel ID | Genotype | Country | Breeding center | Panel ID |
|---|---|---|---|---|---|---|---|
| 126-C409-8-1-2 | COLOMBIA | CIAT | 1 | PCT11\0\0\2\Bo\2\1>181 | COLOMBIA | CIAT | 99 |
| B 22 | BRAZIL | EMBRAPA | 2 | PCT-4 Mad2007\0\1 18-2--1-5-2-3 | COLOMBIA | CIAT | 100 |
| C 537B | MADAGASCAR | FOFIFA-CIRAD | 3 | PCT-4\0\0\1>5-M-1-6 | COLOMBIA | CIAT | 101 |
| C507 1373-1-b-2- - | MADAGASCAR | FOFIFA-CIRAD | 4 | PCT-4\SA\1\1\.SA\2\1>746-1-5-4-1 5-5-1-1-1 | COLOMBIA | CIAT | 102 |
| C630 139-46-2-3-3-b-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 5 | PCT-4\SA\1\1>975-M-2-M-3 2-5-5-1-1 | COLOMBIA | CIAT | 103 |
| C630 38-4-1-b-3-2-1-b-b | MADAGASCAR | FOFIFA-CIRAD | 6 | PCT-4\SA\4\1>1076-2-4-1-5 | COLOMBIA | CIAT | 104 |
| CAIAPO | BRAZIL | 8 | PCT-4\SA\4\1>330-1-4-5-1-M 1-1-1-1-2 | COLOMBIA | CIAT | 105 | |
| CHA LOY OE | THAILAND | 9 | PCT-4\SA\4\1>330-2-2-3-2-M 5-4-4-3-1-5 | COLOMBIA | CIAT | 106 | |
| CHHOMRONG DHAN | NEPAL | 10 | PCT-5\PHB\1\0.PHB\1.PHB\1.PHB\1>78-2--6-2-M | COLOMBIA | CIAT | 107 | |
| CIRAD 141 | BRAZIL | CIRAD | 11 | PRIMAVERA | BRAZIL | IAC | 108 |
| CIRAD 392 | MADAGASCAR | FOFIFA-CIRAD | 12 | SCRID036 4-1-1-5-M | MADAGASCAR | FOFIFA-CIRAD | 109 |
| CIRAD 394 | MADAGASCAR | FOFIFA-CIRAD | 13 | SCRID090 148-1-2-4-5-4-2 | MADAGASCAR | FOFIFA-CIRAD | 110 |
| CIRAD 409 | BRAZIL | CIRAD | 15 | SCRID090 60-1-1-2-4-1-2 | MADAGASCAR | FOFIFA-CIRAD | 111 |
| CIRAD 447 | BRAZIL | CIRAD | 16 | SCRID090 72-3-1-3-5-1-- | MADAGASCAR | FOFIFA-CIRAD | 112 |
| CIRAD 488 | MADAGASCAR | FOFIFA-CIRAD | 17 | SCRID090 89-1-5-4-2-2 | MADAGASCAR | FOFIFA-CIRAD | 113 |
| CNA 4123 | BRAZIL | EMBRAPA | 18 | SCRID091 10-1-3-2-5-3-2 | MADAGASCAR | FOFIFA-CIRAD | 114 |
| CNA 4136 | BRAZIL | EMBRAPA | 19 | SCRID091 11-1-4-3-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 115 |
| CNA 4137 | BRAZIL | EMBRAPA | 20 | SCRID091 15-2-2-1-1-2 | MADAGASCAR | FOFIFA-CIRAD | 116 |
| CNA 4196 | BRAZIL | EMBRAPA | 21 | SCRID091 24-3-2-2-3-5-4 | MADAGASCAR | FOFIFA-CIRAD | 117 |
| CNA-IREM 190 | BRAZIL | EMBRAPA | 22 | SCRID111 1-4-3-3-5-5-4 | MADAGASCAR | FOFIFA-CIRAD | 118 |
| CT13582-15-5-M | COLOMBIA | CIAT | 23 | SCRID128 1-3-4-2-4-4 | MADAGASCAR | FOFIFA-CIRAD | 119 |
| CUIABANA | BRAZIL | 24 | SCRID128 18-5-4-4-5-3 | MADAGASCAR | FOFIFA-CIRAD | 120 | |
| CURINCA | BRAZIL | 25 | SCRID128 21-3-1-1-1-3 | MADAGASCAR | FOFIFA-CIRAD | 122 | |
| DANGREY | BHUTAN | 26 | SCRID136 20-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 123 | |
| DANIELA | BRAZIL | 27 | SCRID139 18-2-4-1-1-3-1 | MADAGASCAR | FOFIFA-CIRAD | 124 | |
| DOURADO PRECOCE | BRAZIL | 28 | SCRID139 9-1-5-2-4-4-1 | MADAGASCAR | FOFIFA-CIRAD | 125 | |
| EARLY MUTANT IAC 165 | BRAZIL | 29 | SCRID195 11-4-4-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 127 | |
| ESTRELA | BRAZIL | 31 | SCRID195 67-1-1-2-2 | MADAGASCAR | FOFIFA-CIRAD | 129 | |
| EXP 003 | MADAGASCAR | FOFIFA-CIRAD | 32 | SCRID195 A1-3-4-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 130 |
| EXP 006 | MADAGASCAR | FOFIFA-CIRAD | 33 | SCRID195-1-5-3 | MADAGASCAR | FOFIFA-CIRAD | 131 |
| EXP 011 | MADAGASCAR | FOFIFA-CIRAD | 34 | SCRID200 15-4-2-4-1 | MADAGASCAR | FOFIFA-CIRAD | 132 |
| EXP 013 | MADAGASCAR | FOFIFA-CIRAD | 35 | SCRID222 122-4-3-3 | MADAGASCAR | FOFIFA-CIRAD | 133 |
| EXP 202 | MADAGASCAR | FOFIFA-CIRAD | 36 | SCRID222 134-1-1-2 | MADAGASCAR | FOFIFA-CIRAD | 134 |
| EXP 206 | MADAGASCAR | FOFIFA-CIRAD | 37 | SCRID222 164-1-1-4 | MADAGASCAR | FOFIFA-CIRAD | 135 |
| EXP 302 | MADAGASCAR | FOFIFA-CIRAD | 38 | SCRID241 1-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 136 |
| EXP 303 | MADAGASCAR | FOFIFA-CIRAD | 39 | SCRID242 22-1-2 | MADAGASCAR | FOFIFA-CIRAD | 137 |
| EXP 304 | MADAGASCAR | FOFIFA-CIRAD | 40 | SCRID243 52-1-1-4 | MADAGASCAR | FOFIFA-CIRAD | 138 |
| EXP 401 | MADAGASCAR | FOFIFA-CIRAD | 41 | SCRID251 25-2-1-2 | MADAGASCAR | FOFIFA-CIRAD | 139 |
| EXP 409 | MADAGASCAR | FOFIFA-CIRAD | 42 | SCRID251 95-1-1-3 | MADAGASCAR | FOFIFA-CIRAD | 140 |
| EXP 910 | MADAGASCAR | FOFIFA-CIRAD | 43 | SCRID252 18-1-2-4 | MADAGASCAR | FOFIFA-CIRAD | 141 |
| F152.06.33.53 13-1-5-1-1 | MADAGASCAR | FOFIFA-CIRAD | 44 | SCRID253 5-2-2-2 | MADAGASCAR | FOFIFA-CIRAD | 142 |
| F154.3G.04.12.10 1 | MADAGASCAR | FOFIFA-CIRAD | 46 | SCRID254 85-3-2-3 | MADAGASCAR | FOFIFA-CIRAD | 143 |
| FOFIFA 116 | MADAGASCAR | FOFIFA-CIRAD | 47 | SCRID260 19-2-1-2 | MADAGASCAR | FOFIFA-CIRAD | 144 |
| FOFIFA 151 | MADAGASCAR | FOFIFA-CIRAD | 48 | SCRID264 69-1-2 | MADAGASCAR | FOFIFA-CIRAD | 145 |
| FOFIFA 159 | MADAGASCAR | FOFIFA-CIRAD | 49 | SCRID271 12-1-3 | MADAGASCAR | FOFIFA-CIRAD | 146 |
| FOFIFA 167 | MADAGASCAR | FOFIFA-CIRAD | 50 | SCRID271 37-1-1 | MADAGASCAR | FOFIFA-CIRAD | 147 |
| FOFIFA 168 | MADAGASCAR | FOFIFA-CIRAD | 51 | SCRID271 67-3-3 | MADAGASCAR | FOFIFA-CIRAD | 148 |
| FOFIFA 171 | MADAGASCAR | FOFIFA-CIRAD | 52 | SCRID273 17-1-2 | MADAGASCAR | FOFIFA-CIRAD | 149 |
| FOFIFA 172 | MADAGASCAR | FOFIFA-CIRAD | 53 | SCRID273 25-1-3 | MADAGASCAR | FOFIFA-CIRAD | 150 |
| FOFIFA 173 | MADAGASCAR | FOFIFA-CIRAD | 54 | SCRID274 30-1-3 | MADAGASCAR | FOFIFA-CIRAD | 151 |
| FOFIFA 180 | MADAGASCAR | FOFIFA-CIRAD | 55 | SCRID275 13-1-5 | MADAGASCAR | FOFIFA-CIRAD | 152 |
| FOFIFA 181 | MADAGASCAR | FOFIFA-CIRAD | 56 | SCRID275 72-5-5 | MADAGASCAR | FOFIFA-CIRAD | 153 |
| FOFIFA 62 | MADAGASCAR | FOFIFA-CIRAD | 57 | SCRID278 148-5-1 | MADAGASCAR | FOFIFA-CIRAD | 154 |
| GUARANI | BRAZIL | 58 | SCRID278 151-5-1 | MADAGASCAR | FOFIFA-CIRAD | 155 | |
| HD 1-4 | France | 59 | SCRID278 42-2-3 | MADAGASCAR | FOFIFA-CIRAD | 156 | |
| IAC 1205 | BRAZIL | IAC | 60 | SCRID292 116-4-2 | MADAGASCAR | FOFIFA-CIRAD | 157 |
| IAC 25 | BRAZIL | IAC | 61 | SCRID292 24-2-5 | MADAGASCAR | FOFIFA-CIRAD | 158 |
| IR53236-275-1 | PHILIPPINES | IRRI | 62 | SCRID6 4-3-M | MADAGASCAR | FOFIFA-CIRAD | 159 |
| IR66421-105-1-1 | PHILIPPINES | IRRI | 63 | SEBOTA 239 | BRAZIL | CIRAD | 160 |
| IRAT 109 | BRAZIL | CIRAD | 64 | SEBOTA 33 | BRAZIL | CIRAD | 161 |
| IRAT 112 | BRAZIL | CIRAD | 65 | SEBOTA 330 | BRAZIL | CIRAD | 162 |
| IRAT 13 | BRAZIL | CIRAD | 66 | SEBOTA 400 | BRAZIL | CIRAD | 164 |
| IRAT 134 | BRAZIL | CIRAD | 67 | SEBOTA 401 | BRAZIL | CIRAD | 165 |
| IRAT 212 | BRAZIL | CIRAD | 68 | SEBOTA 402 | BRAZIL | CIRAD | 166 |
| IRAT 234 | BRAZIL | CIRAD | 69 | SEBOTA 403 | BRAZIL | CIRAD | 167 |
| IRAT 265 | BRAZIL | CIRAD | 70 | SEBOTA 404 | BRAZIL | CIRAD | 168 |
| IRAT 367 | BRAZIL | CIRAD | 71 | SEBOTA 405 | BRAZIL | CIRAD | 169 |
| IRAT 380 | BRAZIL | CIRAD | 72 | SEBOTA 406 | BRAZIL | CIRAD | 170 |
| IREM 239 | BRAZIL | 73 | SEBOTA 408 | BRAZIL | CIRAD | 172 | |
| KUROKA | JAPAN | 74 | SEBOTA 409 | BRAZIL | CIRAD | 173 | |
| LULUWINI 22M | BRAZIL | 75 | SEBOTA 410 | BRAZIL | CIRAD | 174 | |
| MUNUMLIGUERO | BRAZIL | 76 | SEBOTA 65 | BRAZIL | CIRAD | 175 | |
| NABESHI | TAIWAN | 77 | SEBOTA 70 | BRAZIL | CIRAD | 176 | |
| NERICA 1 | IVORY COAST | AFRICARICE | 78 | SEBOTA£ 337 | BRAZIL | CIRAD | 163 |
| NERICA 10 | IVORY COAST | AFRICARICE | 79 | SUCUPIRA | BRAZIL | 177 | |
| NERICA 11 | IVORY COAST | AFRICARICE | 80 | TRES MESES | BRAZIL | 178 | |
| NERICA 12 | IVORY COAST | AFRICARICE | 81 | WAB 450-11-1-P28-1-HB | IVORY COAST | AFRICARICE | 179 |
| NERICA 13 | IVORY COAST | AFRICARICE | 82 | WAB 450-25-2-9-4-1-B-HB | IVORY COAST | AFRICARICE | 180 |
| NERICA 16 | IVORY COAST | AFRICARICE | 83 | WAB 56-125 | IVORY COAST | AFRICARICE | 181 |
| NERICA 18 | IVORY COAST | AFRICARICE | 84 | WAB 56-50 | IVORY COAST | AFRICARICE | 182 |
| NERICA 2 | IVORY COAST | AFRICARICE | 85 | WAB 706-3-4-K4-KB-1 | IVORY COAST | AFRICARICE | 183 |
| NERICA 3 | IVORY COAST | AFRICARICE | 86 | Wab 758 1-1-HB-4 | IVORY COAST | AFRICARICE | 184 |
| NERICA 4 | IVORY COAST | AFRICARICE | 197 | WAB 759-54-2-3-HB-2B | IVORY COAST | AFRICARICE | 185 |
| NERICA 5 | IVORY COAST | AFRICARICE | 87 | WAB 775-95-2-2-HB-1/CIRAD 409-3 1-2-5-3-1 | IVORY COAST | AFRICARICE | 186 |
| NERICA 6 | IVORY COAST | AFRICARICE | 88 | WAB 788-18-2-2-HB-2/PCT-4\SA\1\1>721-M-2-M-4-M-2-M-5-M-1 | IVORY COAST | AFRICARICE | 187 |
| NERICA 7 | IVORY COAST | AFRICARICE | 89 | WAB 878-6-12-1-1-P1-HB | IVORY COAST | AFRICARICE | 188 |
| NERICA 8 | IVORY COAST | AFRICARICE | 90 | WAB 880-1-32-1-1-P2-HB-1 1-2-2 | IVORY COAST | AFRICARICE | 198 |
| NERICA 9 | IVORY COAST | AFRICARICE | 91 | WAB 891SG26 | IVORY COAST | AFRICARICE | 189 |
| PCT11 MAD2007\0\0 14-1-1-1-3-3-2 | COLOMBIA | CIAT | 92 | WAB 891SG9 | IVORY COAST | AFRICARICE | 190 |
| PCT11 MAD2007\0\0 28-3-3-5-5-5 | COLOMBIA | CIAT | 93 | YANGKUM RED | BHUTAN | 191 | |
| PCT11 MAD2007\0\0 3-3-1-3-2-2-4 | COLOMBIA | CIAT | 94 | YUNLU 48 | CHINA | YAAS | 196 |
| PCT11 MAD2007\0\0 3-5-5-2-1-4-4 | COLOMBIA | CIAT | 95 | YUNLU 50 | CHINA | YAAS | 195 |
| PCT11 MAD2007\0\0 50-1-1-1-5-5-5 | COLOMBIA | CIAT | 96 | YUNLU 64 | CHINA | YAAS | 192 |
| PCT11 x CNA7 42-3-2 | COLOMBIA | CIAT | 97 | YUNLU 65 | CHINA | YAAS | 193 |
| PCT11 x CNA7 73-2-5 | COLOMBIA | CIAT | 98 | YUNLU 7 | CHINA | YAAS | 194 |
Table S3. List of 190 rice accessions used for GWAS.
| Genotype | Country | Breeding center | Panel ID | Genotype | Country | Breeding center | Panel ID |
|---|---|---|---|---|---|---|---|
| 126-C409-8-1-2 | COLOMBIA | CIAT | 1 | PCT11\0\0\2\Bo\2\1>181 | COLOMBIA | CIAT | 99 |
| B 22 | BRAZIL | EMBRAPA | 2 | PCT-4 Mad2007\0\1 18-2--1-5-2-3 | COLOMBIA | CIAT | 100 |
| C 537B | MADAGASCAR | FOFIFA-CIRAD | 3 | PCT-4\0\0\1>5-M-1-6 | COLOMBIA | CIAT | 101 |
| C507 1373-1-b-2- - | MADAGASCAR | FOFIFA-CIRAD | 4 | PCT-4\SA\1\1\.SA\2\1>746-1-5-4-1 5-5-1-1-1 | COLOMBIA | CIAT | 102 |
| C630 139-46-2-3-3-b-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 5 | PCT-4\SA\1\1>975-M-2-M-3 2-5-5-1-1 | COLOMBIA | CIAT | 103 |
| C630 38-4-1-b-3-2-1-b-b | MADAGASCAR | FOFIFA-CIRAD | 6 | PCT-4\SA\4\1>1076-2-4-1-5 | COLOMBIA | CIAT | 104 |
| CAIAPO | BRAZIL | 8 | PCT-4\SA\4\1>330-1-4-5-1-M 1-1-1-1-2 | COLOMBIA | CIAT | 105 | |
| CHA LOY OE | THAILAND | 9 | PCT-4\SA\4\1>330-2-2-3-2-M 5-4-4-3-1-5 | COLOMBIA | CIAT | 106 | |
| CHHOMRONG DHAN | NEPAL | 10 | PCT-5\PHB\1\0.PHB\1.PHB\1.PHB\1>78-2--6-2-M | COLOMBIA | CIAT | 107 | |
| CIRAD 141 | BRAZIL | CIRAD | 11 | PRIMAVERA | BRAZIL | IAC | 108 |
| CIRAD 392 | MADAGASCAR | FOFIFA-CIRAD | 12 | SCRID036 4-1-1-5-M | MADAGASCAR | FOFIFA-CIRAD | 109 |
| CIRAD 394 | MADAGASCAR | FOFIFA-CIRAD | 13 | SCRID090 148-1-2-4-5-4-2 | MADAGASCAR | FOFIFA-CIRAD | 110 |
| CIRAD 409 | BRAZIL | CIRAD | 15 | SCRID090 60-1-1-2-4-1-2 | MADAGASCAR | FOFIFA-CIRAD | 111 |
| CIRAD 447 | BRAZIL | CIRAD | 16 | SCRID090 72-3-1-3-5-1-- | MADAGASCAR | FOFIFA-CIRAD | 112 |
| CIRAD 488 | MADAGASCAR | FOFIFA-CIRAD | 17 | SCRID090 89-1-5-4-2-2 | MADAGASCAR | FOFIFA-CIRAD | 113 |
| CNA 4123 | BRAZIL | EMBRAPA | 18 | SCRID091 10-1-3-2-5-3-2 | MADAGASCAR | FOFIFA-CIRAD | 114 |
| CNA 4136 | BRAZIL | EMBRAPA | 19 | SCRID091 11-1-4-3-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 115 |
| CNA 4137 | BRAZIL | EMBRAPA | 20 | SCRID091 15-2-2-1-1-2 | MADAGASCAR | FOFIFA-CIRAD | 116 |
| CNA 4196 | BRAZIL | EMBRAPA | 21 | SCRID091 24-3-2-2-3-5-4 | MADAGASCAR | FOFIFA-CIRAD | 117 |
| CNA-IREM 190 | BRAZIL | EMBRAPA | 22 | SCRID111 1-4-3-3-5-5-4 | MADAGASCAR | FOFIFA-CIRAD | 118 |
| CT13582-15-5-M | COLOMBIA | CIAT | 23 | SCRID128 1-3-4-2-4-4 | MADAGASCAR | FOFIFA-CIRAD | 119 |
| CUIABANA | BRAZIL | 24 | SCRID128 18-5-4-4-5-3 | MADAGASCAR | FOFIFA-CIRAD | 120 | |
| CURINCA | BRAZIL | 25 | SCRID128 21-3-1-1-1-3 | MADAGASCAR | FOFIFA-CIRAD | 122 | |
| DANGREY | BHUTAN | 26 | SCRID136 20-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 123 | |
| DANIELA | BRAZIL | 27 | SCRID139 18-2-4-1-1-3-1 | MADAGASCAR | FOFIFA-CIRAD | 124 | |
| DOURADO PRECOCE | BRAZIL | 28 | SCRID139 9-1-5-2-4-4-1 | MADAGASCAR | FOFIFA-CIRAD | 125 | |
| EARLY MUTANT IAC 165 | BRAZIL | 29 | SCRID195 11-4-4-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 127 | |
| ESTRELA | BRAZIL | 31 | SCRID195 67-1-1-2-2 | MADAGASCAR | FOFIFA-CIRAD | 129 | |
| EXP 003 | MADAGASCAR | FOFIFA-CIRAD | 32 | SCRID195 A1-3-4-2-4-3 | MADAGASCAR | FOFIFA-CIRAD | 130 |
| EXP 006 | MADAGASCAR | FOFIFA-CIRAD | 33 | SCRID195-1-5-3 | MADAGASCAR | FOFIFA-CIRAD | 131 |
| EXP 011 | MADAGASCAR | FOFIFA-CIRAD | 34 | SCRID200 15-4-2-4-1 | MADAGASCAR | FOFIFA-CIRAD | 132 |
| EXP 013 | MADAGASCAR | FOFIFA-CIRAD | 35 | SCRID222 122-4-3-3 | MADAGASCAR | FOFIFA-CIRAD | 133 |
| EXP 202 | MADAGASCAR | FOFIFA-CIRAD | 36 | SCRID222 134-1-1-2 | MADAGASCAR | FOFIFA-CIRAD | 134 |
| EXP 206 | MADAGASCAR | FOFIFA-CIRAD | 37 | SCRID222 164-1-1-4 | MADAGASCAR | FOFIFA-CIRAD | 135 |
| EXP 302 | MADAGASCAR | FOFIFA-CIRAD | 38 | SCRID241 1-1-1-1 | MADAGASCAR | FOFIFA-CIRAD | 136 |
| EXP 303 | MADAGASCAR | FOFIFA-CIRAD | 39 | SCRID242 22-1-2 | MADAGASCAR | FOFIFA-CIRAD | 137 |
| EXP 304 | MADAGASCAR | FOFIFA-CIRAD | 40 | SCRID243 52-1-1-4 | MADAGASCAR | FOFIFA-CIRAD | 138 |
| EXP 401 | MADAGASCAR | FOFIFA-CIRAD | 41 | SCRID251 25-2-1-2 | MADAGASCAR | FOFIFA-CIRAD | 139 |
| EXP 409 | MADAGASCAR | FOFIFA-CIRAD | 42 | SCRID251 95-1-1-3 | MADAGASCAR | FOFIFA-CIRAD | 140 |
| EXP 910 | MADAGASCAR | FOFIFA-CIRAD | 43 | SCRID252 18-1-2-4 | MADAGASCAR | FOFIFA-CIRAD | 141 |
| F152.06.33.53 13-1-5-1-1 | MADAGASCAR | FOFIFA-CIRAD | 44 | SCRID253 5-2-2-2 | MADAGASCAR | FOFIFA-CIRAD | 142 |
| F154.3G.04.12.10 1 | MADAGASCAR | FOFIFA-CIRAD | 46 | SCRID254 85-3-2-3 | MADAGASCAR | FOFIFA-CIRAD | 143 |
| FOFIFA 116 | MADAGASCAR | FOFIFA-CIRAD | 47 | SCRID260 19-2-1-2 | MADAGASCAR | FOFIFA-CIRAD | 144 |
| FOFIFA 151 | MADAGASCAR | FOFIFA-CIRAD | 48 | SCRID264 69-1-2 | MADAGASCAR | FOFIFA-CIRAD | 145 |
| FOFIFA 159 | MADAGASCAR | FOFIFA-CIRAD | 49 | SCRID271 12-1-3 | MADAGASCAR | FOFIFA-CIRAD | 146 |
| FOFIFA 167 | MADAGASCAR | FOFIFA-CIRAD | 50 | SCRID271 37-1-1 | MADAGASCAR | FOFIFA-CIRAD | 147 |
| FOFIFA 168 | MADAGASCAR | FOFIFA-CIRAD | 51 | SCRID271 67-3-3 | MADAGASCAR | FOFIFA-CIRAD | 148 |
| FOFIFA 171 | MADAGASCAR | FOFIFA-CIRAD | 52 | SCRID273 17-1-2 | MADAGASCAR | FOFIFA-CIRAD | 149 |
| FOFIFA 172 | MADAGASCAR | FOFIFA-CIRAD | 53 | SCRID273 25-1-3 | MADAGASCAR | FOFIFA-CIRAD | 150 |
| FOFIFA 173 | MADAGASCAR | FOFIFA-CIRAD | 54 | SCRID274 30-1-3 | MADAGASCAR | FOFIFA-CIRAD | 151 |
| FOFIFA 180 | MADAGASCAR | FOFIFA-CIRAD | 55 | SCRID275 13-1-5 | MADAGASCAR | FOFIFA-CIRAD | 152 |
| FOFIFA 181 | MADAGASCAR | FOFIFA-CIRAD | 56 | SCRID275 72-5-5 | MADAGASCAR | FOFIFA-CIRAD | 153 |
| FOFIFA 62 | MADAGASCAR | FOFIFA-CIRAD | 57 | SCRID278 148-5-1 | MADAGASCAR | FOFIFA-CIRAD | 154 |
| GUARANI | BRAZIL | 58 | SCRID278 151-5-1 | MADAGASCAR | FOFIFA-CIRAD | 155 | |
| HD 1-4 | France | 59 | SCRID278 42-2-3 | MADAGASCAR | FOFIFA-CIRAD | 156 | |
| IAC 1205 | BRAZIL | IAC | 60 | SCRID292 116-4-2 | MADAGASCAR | FOFIFA-CIRAD | 157 |
| IAC 25 | BRAZIL | IAC | 61 | SCRID292 24-2-5 | MADAGASCAR | FOFIFA-CIRAD | 158 |
| IR53236-275-1 | PHILIPPINES | IRRI | 62 | SCRID6 4-3-M | MADAGASCAR | FOFIFA-CIRAD | 159 |
| IR66421-105-1-1 | PHILIPPINES | IRRI | 63 | SEBOTA 239 | BRAZIL | CIRAD | 160 |
| IRAT 109 | BRAZIL | CIRAD | 64 | SEBOTA 33 | BRAZIL | CIRAD | 161 |
| IRAT 112 | BRAZIL | CIRAD | 65 | SEBOTA 330 | BRAZIL | CIRAD | 162 |
| IRAT 13 | BRAZIL | CIRAD | 66 | SEBOTA 400 | BRAZIL | CIRAD | 164 |
| IRAT 134 | BRAZIL | CIRAD | 67 | SEBOTA 401 | BRAZIL | CIRAD | 165 |
| IRAT 212 | BRAZIL | CIRAD | 68 | SEBOTA 402 | BRAZIL | CIRAD | 166 |
| IRAT 234 | BRAZIL | CIRAD | 69 | SEBOTA 403 | BRAZIL | CIRAD | 167 |
| IRAT 265 | BRAZIL | CIRAD | 70 | SEBOTA 404 | BRAZIL | CIRAD | 168 |
| IRAT 367 | BRAZIL | CIRAD | 71 | SEBOTA 405 | BRAZIL | CIRAD | 169 |
| IRAT 380 | BRAZIL | CIRAD | 72 | SEBOTA 406 | BRAZIL | CIRAD | 170 |
| IREM 239 | BRAZIL | 73 | SEBOTA 408 | BRAZIL | CIRAD | 172 | |
| KUROKA | JAPAN | 74 | SEBOTA 409 | BRAZIL | CIRAD | 173 | |
| LULUWINI 22M | BRAZIL | 75 | SEBOTA 410 | BRAZIL | CIRAD | 174 | |
| MUNUMLIGUERO | BRAZIL | 76 | SEBOTA 65 | BRAZIL | CIRAD | 175 | |
| NABESHI | TAIWAN | 77 | SEBOTA 70 | BRAZIL | CIRAD | 176 | |
| NERICA 1 | IVORY COAST | AFRICARICE | 78 | SEBOTA£ 337 | BRAZIL | CIRAD | 163 |
| NERICA 10 | IVORY COAST | AFRICARICE | 79 | SUCUPIRA | BRAZIL | 177 | |
| NERICA 11 | IVORY COAST | AFRICARICE | 80 | TRES MESES | BRAZIL | 178 | |
| NERICA 12 | IVORY COAST | AFRICARICE | 81 | WAB 450-11-1-P28-1-HB | IVORY COAST | AFRICARICE | 179 |
| NERICA 13 | IVORY COAST | AFRICARICE | 82 | WAB 450-25-2-9-4-1-B-HB | IVORY COAST | AFRICARICE | 180 |
| NERICA 16 | IVORY COAST | AFRICARICE | 83 | WAB 56-125 | IVORY COAST | AFRICARICE | 181 |
| NERICA 18 | IVORY COAST | AFRICARICE | 84 | WAB 56-50 | IVORY COAST | AFRICARICE | 182 |
| NERICA 2 | IVORY COAST | AFRICARICE | 85 | WAB 706-3-4-K4-KB-1 | IVORY COAST | AFRICARICE | 183 |
| NERICA 3 | IVORY COAST | AFRICARICE | 86 | Wab 758 1-1-HB-4 | IVORY COAST | AFRICARICE | 184 |
| NERICA 4 | IVORY COAST | AFRICARICE | 197 | WAB 759-54-2-3-HB-2B | IVORY COAST | AFRICARICE | 185 |
| NERICA 5 | IVORY COAST | AFRICARICE | 87 | WAB 775-95-2-2-HB-1/CIRAD 409-3 1-2-5-3-1 | IVORY COAST | AFRICARICE | 186 |
| NERICA 6 | IVORY COAST | AFRICARICE | 88 | WAB 788-18-2-2-HB-2/PCT-4\SA\1\1>721-M-2-M-4-M-2-M-5-M-1 | IVORY COAST | AFRICARICE | 187 |
| NERICA 7 | IVORY COAST | AFRICARICE | 89 | WAB 878-6-12-1-1-P1-HB | IVORY COAST | AFRICARICE | 188 |
| NERICA 8 | IVORY COAST | AFRICARICE | 90 | WAB 880-1-32-1-1-P2-HB-1 1-2-2 | IVORY COAST | AFRICARICE | 198 |
| NERICA 9 | IVORY COAST | AFRICARICE | 91 | WAB 891SG26 | IVORY COAST | AFRICARICE | 189 |
| PCT11 MAD2007\0\0 14-1-1-1-3-3-2 | COLOMBIA | CIAT | 92 | WAB 891SG9 | IVORY COAST | AFRICARICE | 190 |
| PCT11 MAD2007\0\0 28-3-3-5-5-5 | COLOMBIA | CIAT | 93 | YANGKUM RED | BHUTAN | 191 | |
| PCT11 MAD2007\0\0 3-3-1-3-2-2-4 | COLOMBIA | CIAT | 94 | YUNLU 48 | CHINA | YAAS | 196 |
| PCT11 MAD2007\0\0 3-5-5-2-1-4-4 | COLOMBIA | CIAT | 95 | YUNLU 50 | CHINA | YAAS | 195 |
| PCT11 MAD2007\0\0 50-1-1-1-5-5-5 | COLOMBIA | CIAT | 96 | YUNLU 64 | CHINA | YAAS | 192 |
| PCT11 x CNA7 42-3-2 | COLOMBIA | CIAT | 97 | YUNLU 65 | CHINA | YAAS | 193 |
| PCT11 x CNA7 73-2-5 | COLOMBIA | CIAT | 98 | YUNLU 7 | CHINA | YAAS | 194 |
| [1] | Angeles-Shim R B, Asano K, Takashi T, Shim J, Kuroha T, Ayano M, Ashikari M. 2012. A wuschel-related homeobox 3B gene,depilous (dep), confers glabrousness of rice leaves and glumes. Rice, 5(1): 28. |
| [2] | Bradbury P J, Zhang Z W, Kroon D E, Casstevens T M, Ramdoss Y, Buckler E S. 2007. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics, 23(19): 2633-2635. |
| [3] | Browning S R, Browning B L. 2007. Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am J Hum Genet, 81(5): 1084-1097. |
| [4] | Cardinale B J, Duffy J E, Gonzalez A, Hooper D U, Perrings C, Venail P, Narwani A, Mace G M, Tilman D, Wardle D A, Kinzig A P, Daily G C, Loreau M, Grace J B, Larigauderie A, Srivastava D S, Naeem S. 2012. Biodiversity loss and its impact on humanity. Nature, 486: 59‒67. |
| [5] | Chen J G, Zhang Y, Tan Y W, Zhang M, Zhu L L, Xu G H, Fan X R. 2016. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2. 1 expression with the OsNAR2.1 promoter. Plant Biotechnol J, 14: 1705-1715. |
| [6] | Cho Y I, Jiang W Z, Chin J H, Piao Z Z, Cho Y G, McCouch S R, Koh H J. 2007. Identified QTL associated with physiological nitrogen use efficiency in rice. Mol Cells, 23(1): 72-79. |
| [7] | Dawson J C, Huggins D R, Jones S S. 2008. Characterizing nitrogen use efficiency in natural and agricultural ecosystems to improve the performance of cereal crops in low-input and organic agricultural systems. Field Crops Res, 107(2): 89-101. |
| [8] | Elshire R J, Glaubitz J C, Sun Q, Poland J A, Kawamoto K, Buckler E S, Mitchell S E. 2011. A robust, simple genotyping- by-sequencing (GBS) approach for high diversity species. PLoS One, 6(5): e19379. |
| [9] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F. 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet, 112(6): 1164-1171. |
| [10] | Fan X R, Tang Z, Tan Y W, Zhang Y, Luo B B, Yang M, Lian X M, Shen Q R, Miller A J, Xu G H. 2016. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA, 113: 7118-7123. |
| [11] | Gachomo E W, Baptiste L J, Kefela T, Saidel W M, Kotchoni S O. 2014. The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biol, 14: 221. |
| [12] | Gao L M, Lu Z F, Ding L, Guo J J, Wang M, Ling N, Guo S W, Shen Q R. 2018. Role of aquaporins in determining carbon and nitrogen status in higher plants. Int J Mol Sci, 19(1): 35. |
| [13] | Garnett T, Plett D, Heuer S, Okamoto M. 2015. Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: Challenges and future directions. Funct Plant Biol, 42(10): 921-941. |
| [14] | Glaubitz J C, Casstevens T M, Lu F, Harriman J, Eshire R J, Sun Q, Buckler E S. 2014. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS One, 9(2): e90346. |
| [15] | Godfray H C J, Beddington J R, Crute I R, Haddad L, Lawrence D, Muir J F, Pretty J, Robinson S, Thomas S M, Toulmin C. 2010. Food security: The challenge of feeding 9 billion people. Science, 327: 812‒818. |
| [16] | Gondro C, van der Werf J, Hayes B. 2013. Methods in Molecular Biology: Genome-Wide Association Studies and Genomic Prediction. Springer: Humana Press: 1019. |
| [17] | Gordon D, Finch S J. 2005. Factors affecting statistical power in the detection of genetic association. J Clin Invest, 115(6): 1408-1418. |
| [18] | Han B, Huang X H. 2013. Sequencing-based genome-wide association study in rice. Curr Opin Plant Biol, 16(2): 133-138. |
| [19] | Hu B, Wang W, Ou S J, Tang J Y, Li H, Che R H, Zhang Z H, Chai X Y, Wang H R, Wang Y Q, Liang C Z, Liu L C, Piao Z Z, Deng Q Y, Deng K, Xu C, Liang Y, Zhang L H, Li L G, Chu C C. 2015. Variation in NRT1. 1B contributes to nitrate-use divergence between rice subspecies. Nat Genet, 47(7): 834-838. |
| [20] | Ishimaru K, Hirose T, Aoki N, Takahashi S, Ono K, Yamamoto S, Wu J Z, Saji S, Baba T, Ugaki M, Matsumoto T, Ohsgugi R. 2001. Antisense expression of a rice sucrose transporter OsSUT1 in rice (Oryza sativa L.). Plant Cell Physiol, 42(10): 1181-1185. |
| [21] | Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoko M. 2004. A rice semi-dwarf gene, tan-ginbozu (d35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol, 54(4): 533-547. |
| [22] | Jiang D, Fang J J, Lou L M, Zhao J F, Yuan S J, Yin L, Sun W, Peng L X, Guo B T, Li X Y. 2015. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS One, 10(2): e0118169. |
| [23] | Kawahara Y, de la Bastide M, Hamilton J P, Kanamori H, McCombie W R, Ouyang S, Schwartz D C, Tanaka T, Wu J Z, Zhou S G, Childs K L, Davidson R M, Lin H N, Quesada- Ocampo L, Vaillancourt B, Sakai H, Lee S S, Kim J, Numa H, Itoh T, Buell C R, Matsumoto T. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice, 6: 4. |
| [24] | Kong Z S, Li M N, Yang W Q, Xu W Y, Xue Y B. 2006. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol, 141(4): 1376-1388. |
| [25] | Lee S, Kim J, Han J J, Han M J, An G. 2004. Functional analyses of the flowering time gene OsMADS50, the putative suppressor of overexpression of CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J, 38(5): 754-764. |
| [26] | Li D J, Yang C H, Li X B, Gan Q, Zhao X F, Zhu L H. 2009. Functional characterization of rice OsDof12. Planta, 229: 1159‒1169. |
| [27] | Li H, Hu B, Chu C C. 2017. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J Exp Bot, 68(10): 2477-2488. |
| [28] | Li J Y, Wang J, Zeigler R S. 2014. The 3000 rice genomes project: New opportunities and challenges for future rice research. GigaScience, 3(1): 8. |
| [29] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F. 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet, 43(12): 1266-1269. |
| [30] | Li Y F, Li M M, Cao G L, Han L Z. 2010. Effects of genetic background on expression of QTL for nitrogen efficiency in irrigated rice and upland rice. Sci Agric Sin, 43(21): 4331-4340. (in Chinese with English abstract) |
| [31] | Liu Z Y, Zhu C S, Jiang Y, Tian Y L, Yu J, An H Z, Tang W J, Sun J, Tang J P, Chen G M, Zhai H Q, Wang C M, Wan J M. 2016. Association mapping and genetic dissection of nitrogen use efficiency-related traits in rice (Oryza sativa L.). Funct Integr Genom, 16(3): 323-333. |
| [32] | Lu Y E, Song Z Y, Lü K, Lian X M, Cai H M. 2012. Molecular characterization, expression and function analysis of the amino acid transporter gene family(OsAATs) in rice. Acta Physiol Plant, 34: 1943‒1962. |
| [33] | Luo J H, Liu H, Zhou T Y, Gu B G, Huang X H, Shangguan Y Y, Zhu J J, Li Y, Zhao Y, Wang Y C, Zhao Q, Wang A H, Wang Z Q, Sang T, Wang Z X, Han B. 2013. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell, 25(9): 3360-3376. |
| [34] | Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann Bot, 105(7): 1141-1157. |
| [35] | Mather K A, Caicedo A L, Polato N R, Olsen K M, McCouch S, Purugganan M D. 2007. The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics, 177: 2223‒2232. |
| [36] | Moll R H, Kamprath E J, Jackson W A. 1982. Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron J, 74: 562‒564. |
| [37] | Nguyen Q N, Lee Y S, Cho L H, Jeong H J, An G, Jung K H. 2015. Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta, 241(3): 603-613. |
| [38] | Obara M, Kajiura M, Fukuta Y, Yano M, Hayashi M, Yamaya T, Sato T. 2001. Mapping of QTL associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J Exp Bot, 52: 1209‒1217. |
| [39] | Ogawa S, Suzuki Y, Yoshizawa R, Kanno K, Makino A. 2011. Effect of individual suppression of RBCS multigene family on Rubisco contents in rice leaves. Plant Cell Environ, 35(3): 546-553. |
| [40] | Price A L, Patterson N J, Plenge R M, Weinblatt M E, Shadick N A, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 38(8): 904-909. |
| [41] | Raboin L M, Randriambololona T, Radanielina T, Ramanantsoanirina A, Ahmadi N, Dusserre J. 2014. Upland rice varieties for smallholder farming in the cold conditions in Madagascar’s tropical highlands. Field Crops Res, 169: 11‒20. |
| [42] | Raboin L M, Ballini E, Tharreau D, Ramanantsoanirina A, Frouin J, Courtois B, Ahmadi N. 2016. Association mapping of resistance to rice blast in upland field conditions. Rice, 9: 59. |
| [43] | Rakotoson T, Dusserre J, Letourmy P, Ramonta I R, Cao T V, Ramanantsoanirina A, Roumet P, Ahmadi N, Raboin L M. 2017. Genetic variability of nitrogen use efficiency in rainfed upland rice. Field Crops Res, 213: 194‒203. |
| [44] | Raun W R, Johnson G V. 1999. Improving nitrogen use efficiency for cereal production. J Agron, 91(3): 357-363. |
| [45] | Risterucci A M, Grivet L, N’goran J A K, Pieretti I, Flament M H, Lanaud C. 2000. A high-density linkage map of Theobroma cacao L. Theor Appl Genet, 101: 948‒955. |
| [46] | Schmidt R, Schippers J H M, Mieulet D, Obata T, Fernie A R, Guiderdoni E, Mueller-Roeber B. 2013. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J, 76(2): 258-273. |
| [47] | Senthilvel S, Vinod K K, Malarvizhi P, Maheswaran M. 2008. QTL and QTL × environment effects on agronomic and nitrogen acquisition traits in rice. J Integr Plant Biol, 50(9): 1108-1117. |
| [48] | Shan Y H, Wang Y L, Pan X B. 2005. Mapping of QTL for nitrogen use efficiency and related traits in rice (Oryza sativa L.). Agir Sci China, 10: 721‒727. |
| [49] | Shin J H, Blay S, McNeney B, Graham J. 2006. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw, 16(3): 1-10. |
| [50] | Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet, 40(8): 1023-1028. |
| [51] | Storey J D, Tibshirani R. 2003. Statistical significance for genome- wide studies. Proc Natl Acad Sci USA, 100(16): 9440-9445. |
| [52] | Tang W J, Ye J, Yao X M, Zhao P Z, Xuan W, Tian Y L, Zhang Y Y, Xu S, An H Z, Chen G M, Yu J, Wu W, Ge Y W, Liu X L, Li J, Zhang H Z, Zhao Y Q, Yang B, Jiang X Z, Peng C, Zhou C, Terzaghi W, Wang C M, Wan J M. 2019. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat Commun, 10(1): 5279. |
| [53] | Tang Z, Fan X R, Li Q, Feng H M, Miller A J, Shen Q R, Xu G H. 2012. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol, 160(4): 2052‒2063. |
| [54] | Tester M, Langridge P. 2010. Breeding technologies to increase crop production in a changing world. Science, 327: 818‒822. |
| [55] | Wei D, Cui K H, Pan J F, Ye G Y, Xiang J, Nie L X, Huang J L. 2011. Genetic dissection of grain nitrogen use efficiency and grain yield and their relationship in rice. Field Crops Res, 124(3): 340-346. |
| [56] | Yan M, Fan X R, Feng H M, Miller A J, Shen Q R, Xu G H. 2011. Rice OsNAR2. 1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ, 34(8): 1360-1372. |
| [57] | Yang J L, Lee S, Hang R L, Kim S R, Lee Y S, Cao X F, Amasino R, An G. 2013. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J, 73(4): 566-578. |
| [58] | Yano K, Yamamoto E, Aya K, Takeuchi H, Lo P C, Yamasaki M, Yoshida S, Kitano H, Hirano K, Matsuoka M. 2016. Genome wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet, 48: 927‒934. |
| [59] | Yu J M, Pressoir G, Briggs W H, Bi I V, Yamasaki M, Doebley J F, McMullen M D, Gaut B S, Nielsen D M, Holland J B, Kresovich S, Buckler E S. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet, 38(2): 203-208. |
| [60] | Zhang P, Zhong K Z, Qasim S M, Tong H. 2016. Association analysis in rice: From application to utilization. Front Plant Sci, 7: 1202. |
| [61] | Zhang S W, Li C H, Cap J, Zhang Y C, Zhang S Q, Xia Y F, Sun D Y, Sun Y. 2009. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol, 151: 1889‒1901. |
| [62] | Zhang Z, Ersoz E, Lai C Q, Todhunter R J, Tiwari H K, Gore M A, Bradbury P J, Yu J, Arnett D K, Ordovas J M, Buckler E S. 2010. Mixed linear model approach adapted for genome-wide association studies. Nat Genet, 42: 355‒360. |
| [63] | Zhou Y, Tao Y J, Tang D N, Wang J, Zhong J, Wang Y, Yuan Q M, Yu X F, Zhang Y, Wang Y L, Liang G H, Dong G C. 2017. Identification of QTL associated with nitrogen uptake and nitrogen use efficiency using high throughput genotyped CSSLS in rice (Oryza sativa L.). Front Plant Sci, 8: 1166. |
| [64] | Zhu C S, Gore M, Buckler E S, Yu J. 2008. Status and prospects of association mapping in plants. Plant Genome, 1: 5‒20. |
| [65] | Zifarelli G, Pusch M. 2010. CLC transport proteins in plants. FEBS Lett, 584(10): 2122‒2127. |
| [1] | Jiang Hongzhen, Wang Yamei, Lai Liuru, Liu Xintong, Miao Changjian, Liu Ruifang, Li Xiaoyun, Tan Jinfang, Gao Zhenyu, Chen Jingguang. OsAMT1.1 Expression by Nitrate-Inducible Promoter of OsNAR2.1 Increases Nitrogen Use Efficiency and Rice Yield [J]. Rice Science, 2023, 30(3): 222-234. |
| [2] | Amrit Kumar Nayak, Anilkumar C, Sasmita Behera, Rameswar Prasad Sah, Gera Roopa Lavanya, Awadhesh Kumar, Lambodar Behera, Muhammed Azharudheen Tp. Genetic Dissection of Grain Size Traits Through Genome-Wide Association Study Based on Genic Markers in Rice [J]. Rice Science, 2022, 29(5): 462-472. |
| [3] | Oyebanji O. Alagbo, Oluyemisi A. Akinyemiju, Bhagirath S. Chauhan. Weed Management in Rainfed Upland Rice Fields under Varied Agro-Ecologies in Nigeria [J]. Rice Science, 2022, 29(4): 328-339. |
| [4] | Li Lin, Zhang Zheng, Tian Hua, Umair Ashraf, Yousef Alhaj Hamoud, Al Aasmi Alaa, Tang Xiangru, Duan Meiyang, Wang Zaiman, Pan Shenggang. Nitrogen Deep Placement Combined with Straw Mulch Cultivation Enhances Physiological Traits, Grain Yield and Nitrogen Use Efficiency in Mechanical Pot-Seedling Transplanting Rice [J]. Rice Science, 2022, 29(1): 89-100. |
| [5] | Balija Vishalakshi, Bangale Umakanth, Ponnuvel Senguttuvel, Makarand Barbadikar Kalyani, Prasad Madamshetty Srinivas, Rao Durbha Sanjeeva, Yadla Hari, Madhav Maganti Sheshu. Improvement of Upland Rice Variety by Pyramiding Drought Tolerance QTL with Two Major Blast Resistance Genes for Sustainable Rice Production [J]. Rice Science, 2021, 28(5): 493-500. |
| [6] | Panigrahy Madhusmita, Das Subhashree, Poli Yugandhar, Kumar Sahoo Pratap, Kumari Khushbu, C. S. Panigrahi Kishore. Carbon Nanoparticle Exerts Positive Growth Effects with Increase in Productivity by Down-Regulating Phytochrome B and Enhancing Internal Temperature in Rice [J]. Rice Science, 2021, 28(3): 289-300. |
| [7] | Kanta Gaihre Yam, Singh Upendra, D. Bible Wendie, Fugice Jr Job, Sanabria Joaquin. Mitigating N2O and NO Emissions from Direct-Seeded Rice with Nitrification Inhibitor and Urea Deep Placement [J]. Rice Science, 2020, 27(5): 434-444. |
| [8] | Chunmei Xu, Liping Chen, Song Chen, Guang Chu, Danying Wang, Xiufu Zhang. Rhizosphere Aeration Improves Nitrogen Transformation in Soil, and Nitrogen Absorption and Accumulation in Rice Plants [J]. Rice Science, 2020, 27(2): 162-174. |
| [9] | Pandit Elssa, Kumar Panda Rajendra, Sahoo Auromeera, Ranjan Pani Dipti, Kumar Pradhan Sharat. Genetic Relationship and Structure Analysis of Root Growth Angle for Improvement of Drought Avoidance in Early and Mid-Early Maturing Rice Genotypes [J]. Rice Science, 2020, 27(2): 124-132. |
| [10] | Yang Lv, Yueying Wang, Jahan Noushin, Haitao Hu, Ping Chen, Lianguang Shang, Haiyan Lin, Guojun Dong, Jiang Hu, Zhenyu Gao, Qian Qian, Yu Zhang, Longbiao Guo. Genome-Wide Association Analysis and Allelic Mining of Grain Shape-Related Traits in Rice [J]. Rice Science, 2019, 26(6): 384-392. |
| [11] | Zongxiang Chen, Zhiming Feng, Houxiang Kang, Jianhua Zhao, Tianxiao Chen, Qianqian Li, Hongbing Gong, Yafang Zhang, Xijun Chen, Xuebiao Pan, Wende Liu, Guoliang Wang, Shimin Zuo. Identification of New Resistance Loci Against Sheath Blight Disease in Rice Through Genome-Wide Association Study [J]. Rice Science, 2019, 26(1): 21-31. |
| [12] | Mukherjee Mitadru, Padhy Barada, Srinivasan Bharathkumar, Mahadani Pradosh, Yasin Baksh Sk, Donde Ravindra, Nath Singh Onkar, Behera Lambodar, Swain Padmini, Kumar Dash Sushanta. Revealing Genetic Relationship and Prospecting of Novel Donors Among Upland Rice Genotypes Using qDTY-Linked SSR Markers [J]. Rice Science, 2018, 25(6): 308-319. |
| [13] | Lee Jae-Sung, Wissuwa Matthias, B. Zamora Oscar, M. Ismail Abdelbagi. Novel Sources of aus Rice for Zinc Deficiency Tolerance Identified Through Association Analysis Using High-Density SNP Array [J]. Rice Science, 2018, 25(5): 293-296. |
| [14] | Xiongsiyee Vua, Rerkasem Benjavan, Veeradittakit Jeeraporn, Saenchai Chorpet, Lordkaew Sittichai, Thebault Prom-u-thai Chanakan. Variation in Grain Quality of Upland Rice from Luang Prabang Province, Lao PDR [J]. Rice Science, 2018, 25(2): 94-102. |
| [15] | Ya-fang Zhang, Yu-yin Ma, Zong-xiang Chen, Jie Zou, Tian-xiao Chen, Qian-qian Li, Xue-biao Pan, Shi-min Zuo. Genome-Wide Association Studies Reveal New Genetic Targets for Five Panicle Traits of International Rice Varieties [J]. Rice Science, 2015, 22(5): 217-226. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||